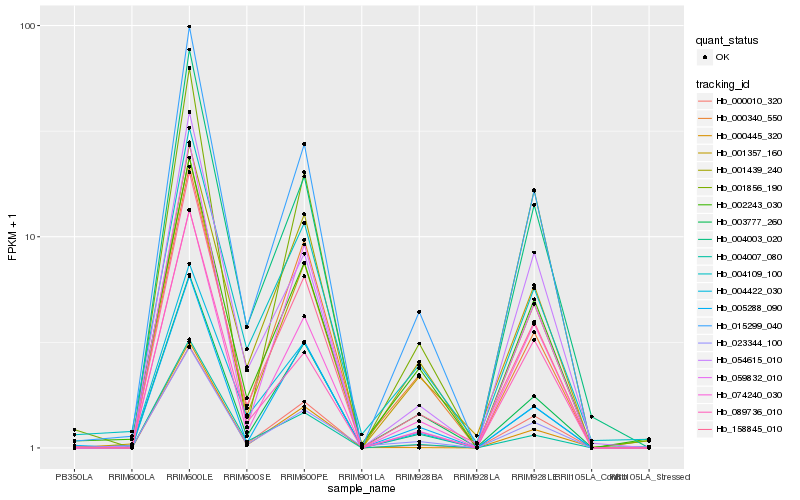
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158845_010 |
0.0 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 [Jatropha curcas] |
| 2 |
Hb_015299_040 |
0.0448481413 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 3 |
Hb_000010_320 |
0.0588490669 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 4 |
Hb_059832_010 |
0.0790505915 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 5 |
Hb_074240_030 |
0.0799834688 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 6 |
Hb_001357_160 |
0.0873216866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_054615_010 |
0.0873866523 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 8 |
Hb_002243_030 |
0.0947803157 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_004003_020 |
0.0953261126 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_004109_100 |
0.1022771784 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 11 |
Hb_004422_030 |
0.1023460359 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK7-like [Jatropha curcas] |
| 12 |
Hb_023344_100 |
0.104247066 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 13 |
Hb_004007_080 |
0.1046980482 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g49900 [Jatropha curcas] |
| 14 |
Hb_000340_550 |
0.1060571159 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 15 |
Hb_001439_240 |
0.1068099673 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 16 |
Hb_089736_010 |
0.1096445195 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |
| 17 |
Hb_003777_260 |
0.1107256435 |
- |
- |
PREDICTED: uncharacterized protein LOC105640927 isoform X4 [Jatropha curcas] |
| 18 |
Hb_005288_090 |
0.110845925 |
- |
- |
PREDICTED: uncharacterized protein LOC105647753 [Jatropha curcas] |
| 19 |
Hb_001856_190 |
0.1116639413 |
- |
- |
SP1L, putative [Ricinus communis] |
| 20 |
Hb_000445_320 |
0.1118649708 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |