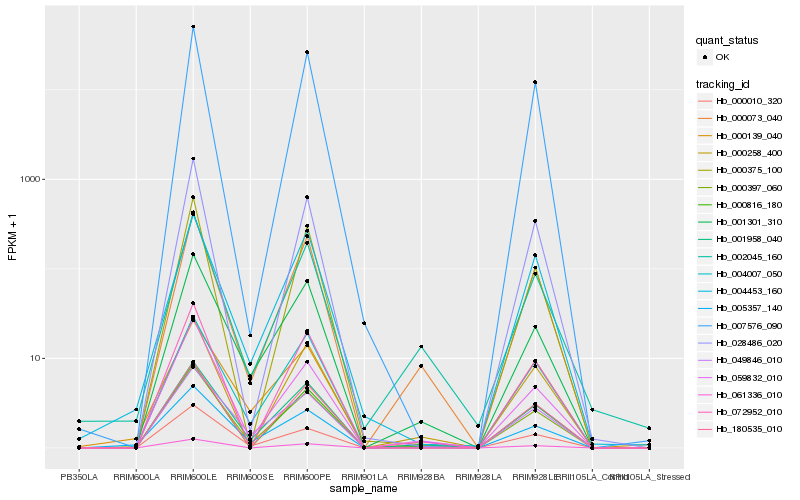
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000397_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 2 |
Hb_001958_040 |
0.0524760321 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 3 |
Hb_000073_040 |
0.0609905883 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 4 |
Hb_005357_140 |
0.071496313 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein A precursor, putative [Ricinus communis] |
| 5 |
Hb_072952_010 |
0.0798884427 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g18430-like [Populus euphratica] |
| 6 |
Hb_000010_320 |
0.0800836157 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 7 |
Hb_001301_310 |
0.0802040337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004007_050 |
0.0808344444 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |
| 9 |
Hb_002045_160 |
0.0852990302 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 10 |
Hb_000258_400 |
0.0885198697 |
transcription factor |
TF Family: bHLH |
transcription regulator, putative [Ricinus communis] |
| 11 |
Hb_061336_010 |
0.0889996983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007576_090 |
0.0894258457 |
- |
- |
lipid transfer protein precursor [Gossypium herbaceum subsp. africanum] |
| 13 |
Hb_049846_010 |
0.090214925 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 14 |
Hb_000816_180 |
0.0917710865 |
- |
- |
PREDICTED: uncharacterized protein LOC105780682 [Gossypium raimondii] |
| 15 |
Hb_180535_010 |
0.0932209389 |
- |
- |
- |
| 16 |
Hb_028486_020 |
0.0935495782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_059832_010 |
0.0937541477 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 18 |
Hb_000139_040 |
0.0947106269 |
- |
- |
spermine synthase, putative [Ricinus communis] |
| 19 |
Hb_004453_160 |
0.096655292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000375_100 |
0.098307978 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |