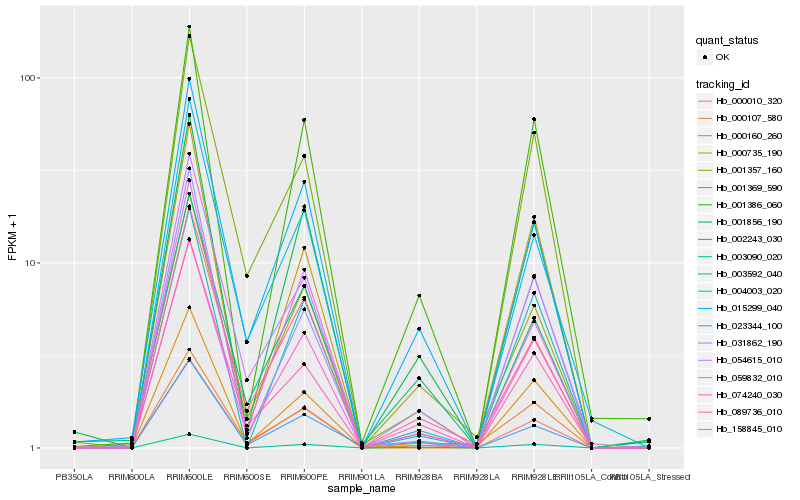
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074240_030 |
0.0 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 2 |
Hb_000010_320 |
0.0567386637 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 3 |
Hb_001856_190 |
0.0649809504 |
- |
- |
SP1L, putative [Ricinus communis] |
| 4 |
Hb_015299_040 |
0.0710342011 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 5 |
Hb_054615_010 |
0.0758151885 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 6 |
Hb_001357_160 |
0.0772905665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003090_020 |
0.0791809079 |
- |
- |
PREDICTED: uncharacterized protein LOC105646662 [Jatropha curcas] |
| 8 |
Hb_158845_010 |
0.0799834688 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 [Jatropha curcas] |
| 9 |
Hb_001386_060 |
0.0820652668 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 10 |
Hb_089736_010 |
0.0905805732 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |
| 11 |
Hb_003592_040 |
0.092895944 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 12 |
Hb_059832_010 |
0.0950638269 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 13 |
Hb_002243_030 |
0.1047824442 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001369_590 |
0.1061800827 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 15 |
Hb_000160_260 |
0.1067446755 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 16 |
Hb_004003_020 |
0.1090621852 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_023344_100 |
0.1102716688 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 18 |
Hb_000735_190 |
0.1144422667 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 19 |
Hb_000107_580 |
0.1164032043 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 20 |
Hb_031862_190 |
0.1168760648 |
- |
- |
- |