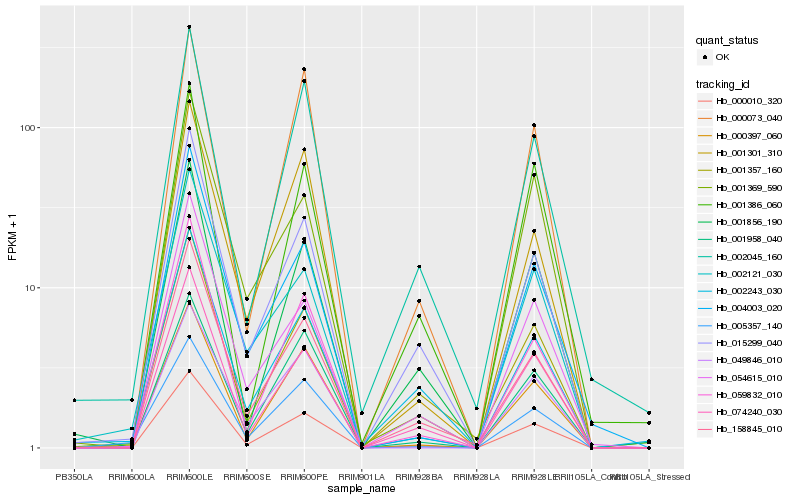
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000010_320 |
0.0 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 2 |
Hb_074240_030 |
0.0567386637 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 3 |
Hb_158845_010 |
0.0588490669 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 [Jatropha curcas] |
| 4 |
Hb_059832_010 |
0.0671266952 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 5 |
Hb_015299_040 |
0.068205809 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 6 |
Hb_000397_060 |
0.0800836157 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 7 |
Hb_001856_190 |
0.0879595259 |
- |
- |
SP1L, putative [Ricinus communis] |
| 8 |
Hb_004003_020 |
0.0881737861 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_054615_010 |
0.0890059636 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 10 |
Hb_001958_040 |
0.0913914808 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 11 |
Hb_001357_160 |
0.0955264014 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002045_160 |
0.0980963369 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 13 |
Hb_000073_040 |
0.101188257 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 14 |
Hb_001301_310 |
0.1035608049 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_049846_010 |
0.1041620894 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 16 |
Hb_005357_140 |
0.1046564021 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein A precursor, putative [Ricinus communis] |
| 17 |
Hb_001386_060 |
0.1082004093 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 18 |
Hb_001369_590 |
0.1094717785 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 19 |
Hb_002121_030 |
0.1096485482 |
- |
- |
Ribonuclease P protein subunit P38-related isoform 1 [Theobroma cacao] |
| 20 |
Hb_002243_030 |
0.1106419251 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |