| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
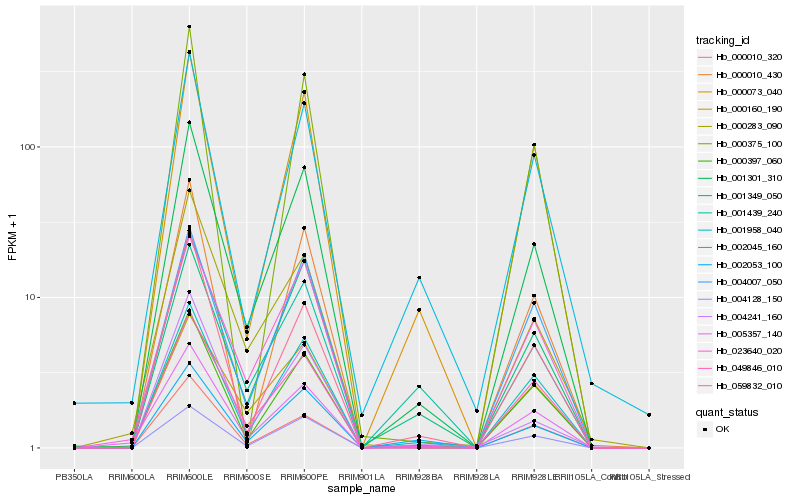
Hb_001301_310 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001958_040 |
0.0700796409 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 3 |
Hb_000397_060 |
0.0802040337 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 4 |
Hb_004128_150 |
0.0868110742 |
- |
- |
hypothetical protein POPTR_0016s08360g [Populus trichocarpa] |
| 5 |
Hb_002053_100 |
0.0868688024 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 6 |
Hb_000073_040 |
0.0916564028 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 7 |
Hb_005357_140 |
0.093332548 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein A precursor, putative [Ricinus communis] |
| 8 |
Hb_049846_010 |
0.0954505365 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 9 |
Hb_023640_020 |
0.0964399034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004007_050 |
0.0991158379 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |
| 11 |
Hb_000010_320 |
0.1035608049 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 12 |
Hb_000283_090 |
0.1049004584 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_059832_010 |
0.1063010189 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 14 |
Hb_001349_050 |
0.1066234385 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 15 |
Hb_001439_240 |
0.1075145269 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 16 |
Hb_000160_190 |
0.1085056215 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 17 |
Hb_002045_160 |
0.111544621 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 18 |
Hb_000010_430 |
0.112421067 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 19 |
Hb_004241_160 |
0.1129483589 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-7 [Jatropha curcas] |
| 20 |
Hb_000375_100 |
0.1132053141 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |