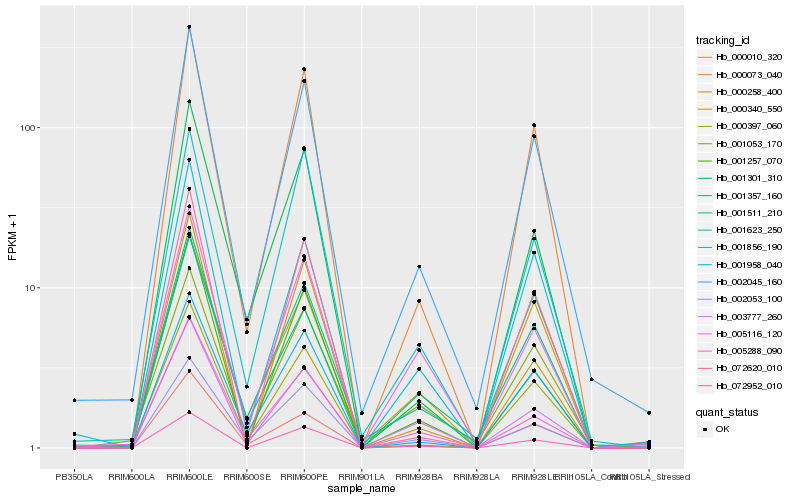
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002045_160 |
0.0 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 2 |
Hb_000073_040 |
0.0781667179 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 3 |
Hb_001053_170 |
0.0838253998 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 4 |
Hb_000397_060 |
0.0852990302 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 5 |
Hb_001958_040 |
0.0920979886 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 6 |
Hb_072620_010 |
0.092824106 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Cucumis melo] |
| 7 |
Hb_000010_320 |
0.0980963369 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 8 |
Hb_000258_400 |
0.0991342649 |
transcription factor |
TF Family: bHLH |
transcription regulator, putative [Ricinus communis] |
| 9 |
Hb_001856_190 |
0.1008240349 |
- |
- |
SP1L, putative [Ricinus communis] |
| 10 |
Hb_000340_550 |
0.101119505 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 11 |
Hb_003777_260 |
0.1032038231 |
- |
- |
PREDICTED: uncharacterized protein LOC105640927 isoform X4 [Jatropha curcas] |
| 12 |
Hb_002053_100 |
0.1047259683 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 13 |
Hb_005288_090 |
0.1060211831 |
- |
- |
PREDICTED: uncharacterized protein LOC105647753 [Jatropha curcas] |
| 14 |
Hb_001257_070 |
0.1067884172 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1-like [Jatropha curcas] |
| 15 |
Hb_001623_250 |
0.1070785683 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 16 |
Hb_005116_120 |
0.107835811 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 17 |
Hb_072952_010 |
0.1095372888 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g18430-like [Populus euphratica] |
| 18 |
Hb_001511_210 |
0.1111657869 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 19 |
Hb_001301_310 |
0.111544621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001357_160 |
0.111602775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |