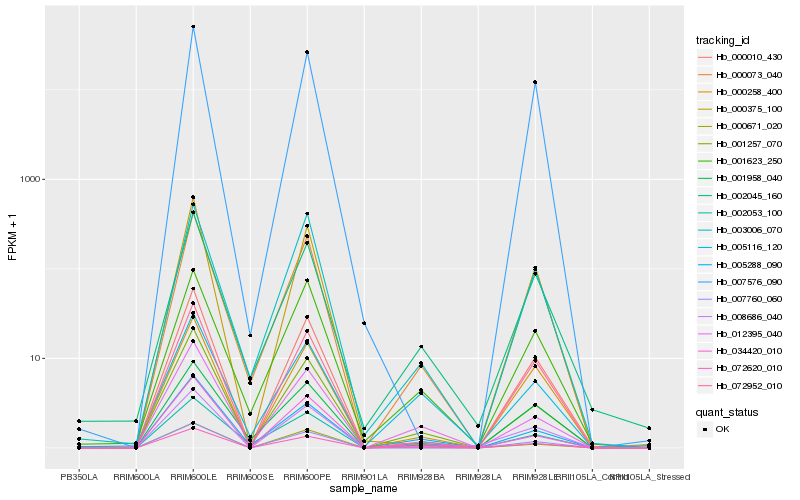
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_072620_010 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Cucumis melo] |
| 2 |
Hb_000073_040 |
0.0767020275 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 3 |
Hb_000258_400 |
0.0808458927 |
transcription factor |
TF Family: bHLH |
transcription regulator, putative [Ricinus communis] |
| 4 |
Hb_072952_010 |
0.0847143253 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g18430-like [Populus euphratica] |
| 5 |
Hb_001257_070 |
0.0882479298 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1-like [Jatropha curcas] |
| 6 |
Hb_000671_020 |
0.0884544004 |
- |
- |
PREDICTED: RNA-binding protein 38 [Jatropha curcas] |
| 7 |
Hb_002045_160 |
0.092824106 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 8 |
Hb_008686_040 |
0.0995040752 |
- |
- |
PREDICTED: uncharacterized protein LOC105632278 isoform X3 [Jatropha curcas] |
| 9 |
Hb_001623_250 |
0.1010985616 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 10 |
Hb_012395_040 |
0.1012448694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000375_100 |
0.1050587123 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 12 |
Hb_002053_100 |
0.1059520577 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 13 |
Hb_005288_090 |
0.1060592328 |
- |
- |
PREDICTED: uncharacterized protein LOC105647753 [Jatropha curcas] |
| 14 |
Hb_007760_060 |
0.1063878536 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0010s17470g [Populus trichocarpa] |
| 15 |
Hb_034420_010 |
0.107559926 |
- |
- |
glutathione S-transferase [Caragana korshinskii] |
| 16 |
Hb_000010_430 |
0.1079824823 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 17 |
Hb_003006_070 |
0.1097092718 |
- |
- |
aquaporin [Manihot esculenta] |
| 18 |
Hb_007576_090 |
0.111747226 |
- |
- |
lipid transfer protein precursor [Gossypium herbaceum subsp. africanum] |
| 19 |
Hb_001958_040 |
0.1124289434 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 20 |
Hb_005116_120 |
0.1125132141 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |