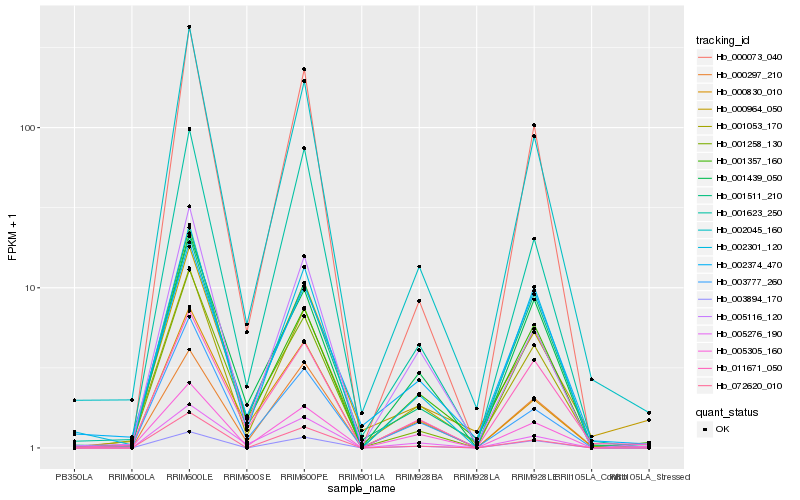
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001053_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 2 |
Hb_002045_160 |
0.0838253998 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 3 |
Hb_001511_210 |
0.0960418852 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 4 |
Hb_002301_120 |
0.1056894947 |
- |
- |
PREDICTED: uncharacterized protein LOC105649581 [Jatropha curcas] |
| 5 |
Hb_011671_050 |
0.1093221834 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 6 |
Hb_005116_120 |
0.1096319069 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001439_050 |
0.1098603051 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 8 |
Hb_000964_050 |
0.1140910532 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 9 |
Hb_000073_040 |
0.1144227885 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 10 |
Hb_001258_130 |
0.1157056187 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 11 |
Hb_005305_160 |
0.1163808496 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002374_470 |
0.1173727734 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_003777_260 |
0.1181752779 |
- |
- |
PREDICTED: uncharacterized protein LOC105640927 isoform X4 [Jatropha curcas] |
| 14 |
Hb_000830_010 |
0.1199695634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005276_190 |
0.1205922765 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000297_210 |
0.1211177505 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 17 |
Hb_003894_170 |
0.1212094487 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 18 |
Hb_001623_250 |
0.1220104052 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 19 |
Hb_072620_010 |
0.1223069002 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Cucumis melo] |
| 20 |
Hb_001357_160 |
0.1230524131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |