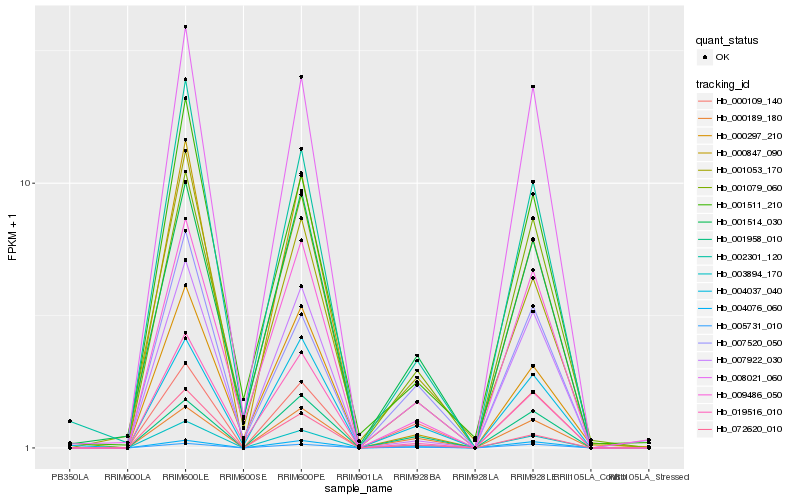
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003894_170 |
0.0 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 2 |
Hb_002301_120 |
0.094711487 |
- |
- |
PREDICTED: uncharacterized protein LOC105649581 [Jatropha curcas] |
| 3 |
Hb_000109_140 |
0.0963943491 |
- |
- |
hypothetical protein CICLE_v100314482mg, partial [Citrus clementina] |
| 4 |
Hb_000189_180 |
0.1034754211 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 5 |
Hb_001079_060 |
0.1124826074 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004037_040 |
0.1130242115 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 7 |
Hb_004076_060 |
0.1148769073 |
- |
- |
PREDICTED: UDP-glycosyltransferase 82A1 [Jatropha curcas] |
| 8 |
Hb_005731_010 |
0.1152389338 |
- |
- |
hypothetical protein VITISV_030941 [Vitis vinifera] |
| 9 |
Hb_000847_090 |
0.1172567312 |
- |
- |
PREDICTED: uncharacterized protein LOC105644263 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001053_170 |
0.1212094487 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 11 |
Hb_001514_030 |
0.1212745436 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 12 |
Hb_001958_010 |
0.1224618008 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 13 |
Hb_007922_030 |
0.124541364 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 14 |
Hb_019516_010 |
0.1265459156 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 15 |
Hb_008021_060 |
0.1268973466 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 16 |
Hb_001511_210 |
0.1271365841 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 17 |
Hb_000297_210 |
0.1274702731 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 18 |
Hb_007520_050 |
0.1279129742 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 19 |
Hb_072620_010 |
0.1281964163 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Cucumis melo] |
| 20 |
Hb_009486_050 |
0.1292948954 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |