| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
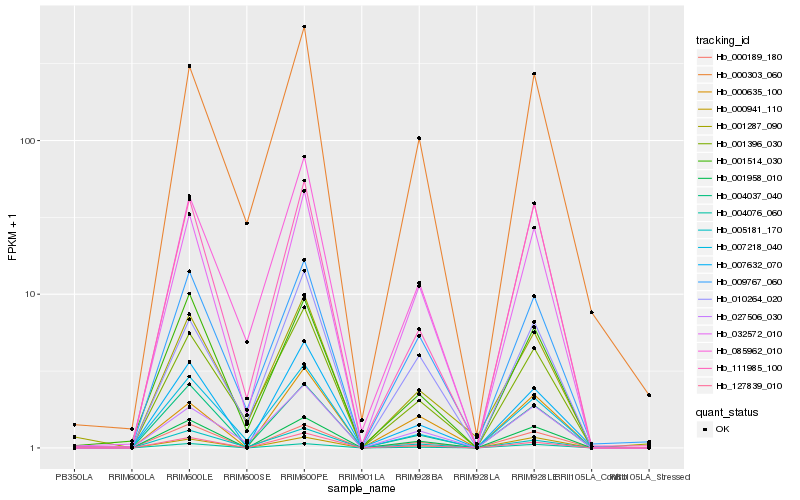
Hb_032572_010 |
0.0 |
- |
- |
PREDICTED: phospholipase A2-alpha [Jatropha curcas] |
| 2 |
Hb_001958_010 |
0.0809991662 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 3 |
Hb_027506_030 |
0.0813345198 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 4 |
Hb_009767_060 |
0.0823210674 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 5 |
Hb_085962_010 |
0.0911156274 |
- |
- |
hypothetical protein POPTR_0017s11380g [Populus trichocarpa] |
| 6 |
Hb_000189_180 |
0.0939884428 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 7 |
Hb_000303_060 |
0.0946879169 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_010264_020 |
0.0972560344 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_001396_030 |
0.0981041086 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |
| 10 |
Hb_127839_010 |
0.1025299388 |
- |
- |
hypothetical protein PRUPE_ppa000956mg [Prunus persica] |
| 11 |
Hb_111985_100 |
0.1050531525 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 12 |
Hb_004037_040 |
0.1082405437 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 13 |
Hb_000635_100 |
0.111213048 |
- |
- |
- |
| 14 |
Hb_007632_070 |
0.1117592171 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_004076_060 |
0.1137832763 |
- |
- |
PREDICTED: UDP-glycosyltransferase 82A1 [Jatropha curcas] |
| 16 |
Hb_005181_170 |
0.1150715977 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |
| 17 |
Hb_000941_110 |
0.1150908307 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 18 |
Hb_001514_030 |
0.1153117529 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 19 |
Hb_007218_040 |
0.1185104565 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 20 |
Hb_001287_090 |
0.1190547519 |
- |
- |
Endoglucanase 4 [Aegilops tauschii] |