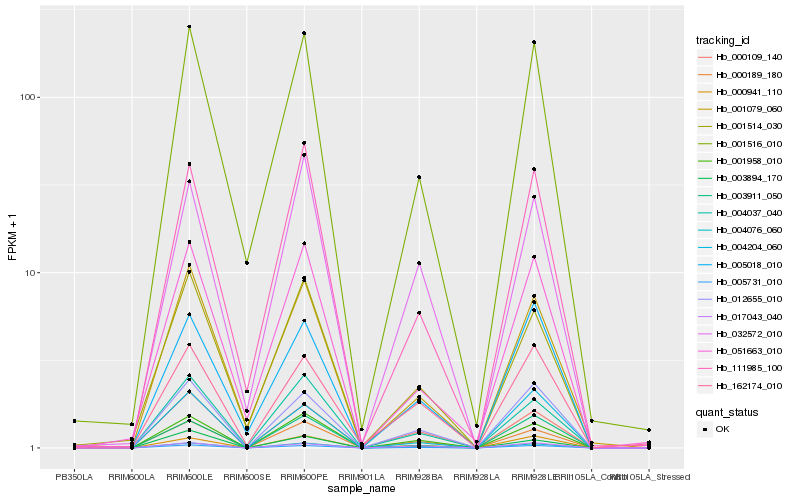
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004076_060 |
0.0 |
- |
- |
PREDICTED: UDP-glycosyltransferase 82A1 [Jatropha curcas] |
| 2 |
Hb_005731_010 |
0.0398969261 |
- |
- |
hypothetical protein VITISV_030941 [Vitis vinifera] |
| 3 |
Hb_001958_010 |
0.0483378681 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 4 |
Hb_000189_180 |
0.0532545315 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 5 |
Hb_012655_010 |
0.0678381376 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 6 |
Hb_000941_110 |
0.0679777245 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 7 |
Hb_000109_140 |
0.0860980498 |
- |
- |
hypothetical protein CICLE_v100314482mg, partial [Citrus clementina] |
| 8 |
Hb_001516_010 |
0.0937163085 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 9 |
Hb_111985_100 |
0.0940757645 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 10 |
Hb_004037_040 |
0.1002466072 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 11 |
Hb_003911_050 |
0.1027599818 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 12 |
Hb_001079_060 |
0.1056219255 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_051663_010 |
0.1087431169 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 14 |
Hb_032572_010 |
0.1137832763 |
- |
- |
PREDICTED: phospholipase A2-alpha [Jatropha curcas] |
| 15 |
Hb_003894_170 |
0.1148769073 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 16 |
Hb_001514_030 |
0.1156211557 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 17 |
Hb_017043_040 |
0.1158697397 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 18 |
Hb_005018_010 |
0.1166308596 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 19 |
Hb_004204_060 |
0.117686781 |
transcription factor |
TF Family: NAC |
NAC transcription factor 058, partial [Jatropha curcas] |
| 20 |
Hb_162174_010 |
0.1208622551 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |