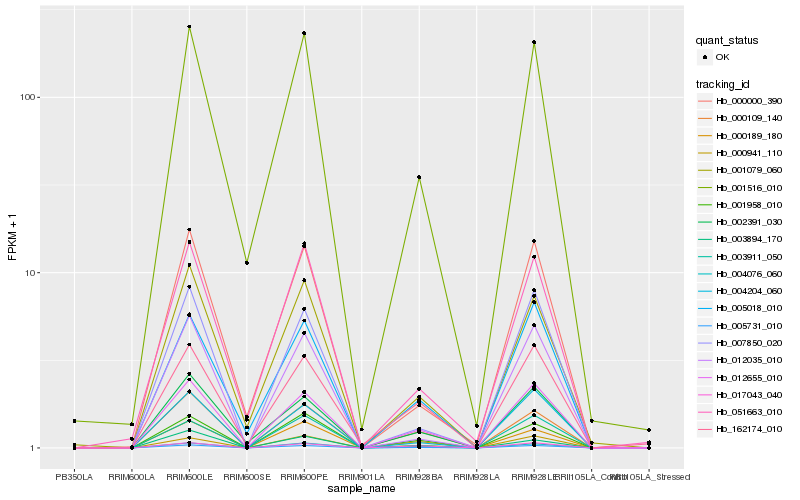
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005731_010 |
0.0 |
- |
- |
hypothetical protein VITISV_030941 [Vitis vinifera] |
| 2 |
Hb_012655_010 |
0.0320909443 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 3 |
Hb_004076_060 |
0.0398969261 |
- |
- |
PREDICTED: UDP-glycosyltransferase 82A1 [Jatropha curcas] |
| 4 |
Hb_000189_180 |
0.0757607958 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 5 |
Hb_000109_140 |
0.0790871971 |
- |
- |
hypothetical protein CICLE_v100314482mg, partial [Citrus clementina] |
| 6 |
Hb_000941_110 |
0.0802835795 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 7 |
Hb_001958_010 |
0.0820216246 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 8 |
Hb_004204_060 |
0.0954727997 |
transcription factor |
TF Family: NAC |
NAC transcription factor 058, partial [Jatropha curcas] |
| 9 |
Hb_001516_010 |
0.0980218172 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 10 |
Hb_017043_040 |
0.100913667 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 11 |
Hb_002391_030 |
0.1035157781 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 12 |
Hb_005018_010 |
0.1041306142 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 13 |
Hb_007850_020 |
0.1059280289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_162174_010 |
0.106479535 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_001079_060 |
0.1096113168 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003911_050 |
0.1144083036 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 17 |
Hb_003894_170 |
0.1152389338 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 18 |
Hb_012035_010 |
0.1156181638 |
- |
- |
hypothetical protein JCGZ_21143 [Jatropha curcas] |
| 19 |
Hb_051663_010 |
0.1188231981 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 20 |
Hb_000000_390 |
0.1206620419 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |