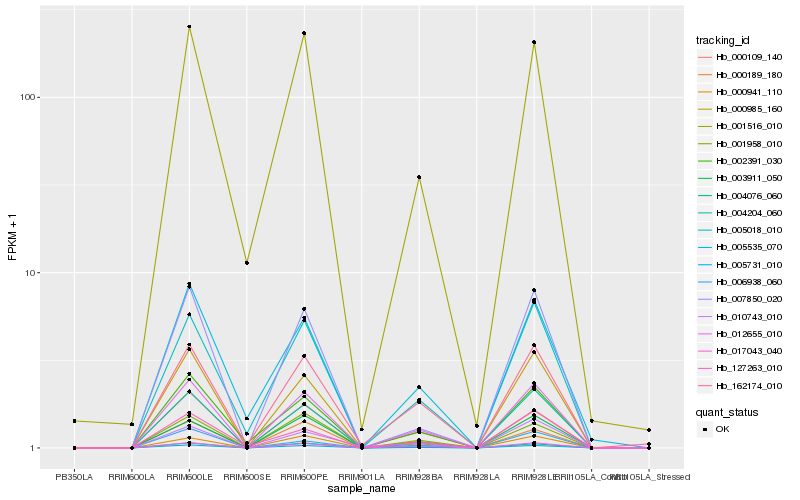
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012655_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 2 |
Hb_005731_010 |
0.0320909443 |
- |
- |
hypothetical protein VITISV_030941 [Vitis vinifera] |
| 3 |
Hb_004076_060 |
0.0678381376 |
- |
- |
PREDICTED: UDP-glycosyltransferase 82A1 [Jatropha curcas] |
| 4 |
Hb_000941_110 |
0.0835114452 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 5 |
Hb_017043_040 |
0.0837774163 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 6 |
Hb_000109_140 |
0.0903866189 |
- |
- |
hypothetical protein CICLE_v100314482mg, partial [Citrus clementina] |
| 7 |
Hb_004204_060 |
0.0932371071 |
transcription factor |
TF Family: NAC |
NAC transcription factor 058, partial [Jatropha curcas] |
| 8 |
Hb_162174_010 |
0.0949118401 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_005018_010 |
0.0952421109 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_000189_180 |
0.0993008001 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 11 |
Hb_002391_030 |
0.1009308685 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 12 |
Hb_001958_010 |
0.1061560517 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 13 |
Hb_001516_010 |
0.1116356413 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 14 |
Hb_007850_020 |
0.1126355122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_010743_010 |
0.1254088674 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_003911_050 |
0.1267557203 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 17 |
Hb_000985_160 |
0.1270668877 |
- |
- |
PREDICTED: cytochrome P450 724B1-like [Jatropha curcas] |
| 18 |
Hb_005535_070 |
0.1278731789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_127263_010 |
0.1285245932 |
- |
- |
hypothetical protein JCGZ_02828 [Jatropha curcas] |
| 20 |
Hb_006938_060 |
0.1292050602 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |