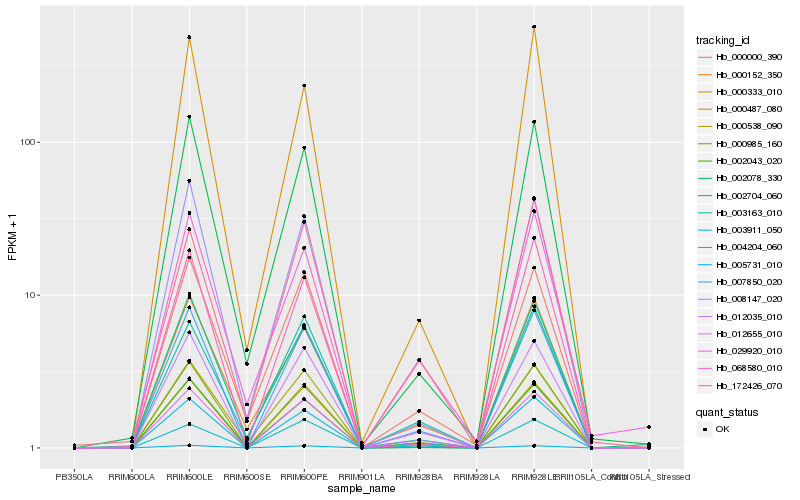
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004204_060 |
0.0 |
transcription factor |
TF Family: NAC |
NAC transcription factor 058, partial [Jatropha curcas] |
| 2 |
Hb_007850_020 |
0.0346840848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000985_160 |
0.0434797561 |
- |
- |
PREDICTED: cytochrome P450 724B1-like [Jatropha curcas] |
| 4 |
Hb_012035_010 |
0.0663485857 |
- |
- |
hypothetical protein JCGZ_21143 [Jatropha curcas] |
| 5 |
Hb_000152_350 |
0.0915510988 |
- |
- |
PREDICTED: enolase [Jatropha curcas] |
| 6 |
Hb_012655_010 |
0.0932371071 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 7 |
Hb_002078_330 |
0.0945708633 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 8 |
Hb_003163_010 |
0.0946352382 |
- |
- |
hypothetical protein POPTR_0012s02920g [Populus trichocarpa] |
| 9 |
Hb_005731_010 |
0.0954727997 |
- |
- |
hypothetical protein VITISV_030941 [Vitis vinifera] |
| 10 |
Hb_003911_050 |
0.09646558 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 11 |
Hb_000000_390 |
0.0991674762 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 12 |
Hb_002043_020 |
0.1006503949 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 13 |
Hb_029920_010 |
0.1013073843 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 14 |
Hb_000538_090 |
0.1058288948 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1603380 [Ricinus communis] |
| 15 |
Hb_000487_080 |
0.1059157458 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 16 |
Hb_068580_010 |
0.1063562053 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X2 [Populus euphratica] |
| 17 |
Hb_172426_070 |
0.1074712175 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_008147_020 |
0.108273822 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 19 |
Hb_002704_060 |
0.1099545395 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 20 |
Hb_000333_010 |
0.1109424516 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |