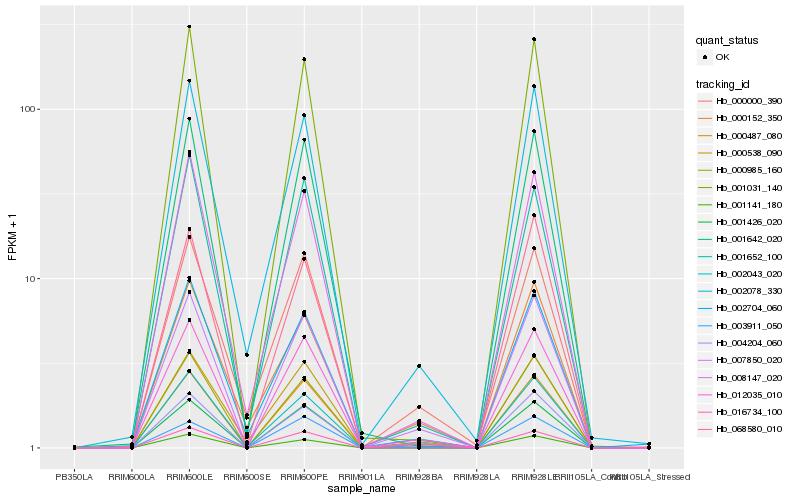
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007850_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000985_160 |
0.0317431268 |
- |
- |
PREDICTED: cytochrome P450 724B1-like [Jatropha curcas] |
| 3 |
Hb_012035_010 |
0.0326764255 |
- |
- |
hypothetical protein JCGZ_21143 [Jatropha curcas] |
| 4 |
Hb_004204_060 |
0.0346840848 |
transcription factor |
TF Family: NAC |
NAC transcription factor 058, partial [Jatropha curcas] |
| 5 |
Hb_000487_080 |
0.0767385755 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 6 |
Hb_002078_330 |
0.0784674742 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 7 |
Hb_002043_020 |
0.079097379 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 8 |
Hb_008147_020 |
0.0831955676 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 9 |
Hb_000538_090 |
0.085709537 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1603380 [Ricinus communis] |
| 10 |
Hb_000000_390 |
0.086860782 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 11 |
Hb_001031_140 |
0.0873816821 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g33370-like [Jatropha curcas] |
| 12 |
Hb_002704_060 |
0.0923827491 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 13 |
Hb_001141_180 |
0.0928075415 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 14 |
Hb_001426_020 |
0.0930355373 |
- |
- |
hypothetical protein CICLE_v10033594mg, partial [Citrus clementina] |
| 15 |
Hb_003911_050 |
0.0934435157 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 16 |
Hb_001642_020 |
0.0948222724 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 17 |
Hb_000152_350 |
0.0948907701 |
- |
- |
PREDICTED: enolase [Jatropha curcas] |
| 18 |
Hb_016734_100 |
0.0963484991 |
- |
- |
- |
| 19 |
Hb_068580_010 |
0.098107531 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X2 [Populus euphratica] |
| 20 |
Hb_001652_100 |
0.1020105311 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |