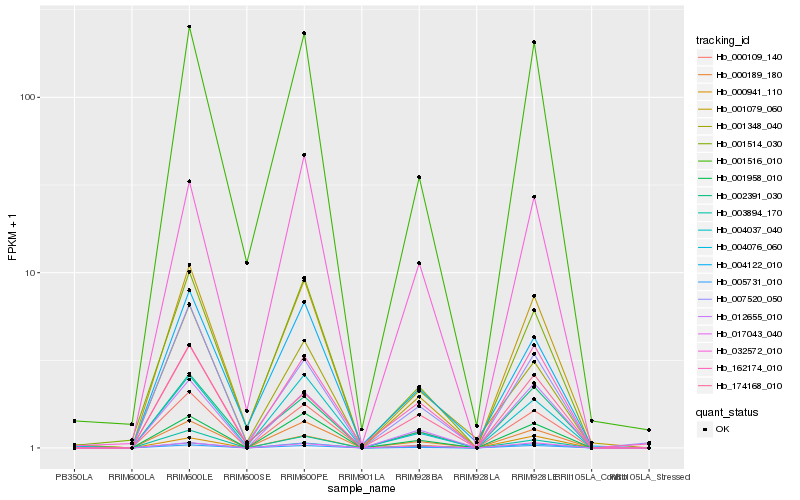
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000109_140 |
0.0 |
- |
- |
hypothetical protein CICLE_v100314482mg, partial [Citrus clementina] |
| 2 |
Hb_000189_180 |
0.0583790827 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 3 |
Hb_005731_010 |
0.0790871971 |
- |
- |
hypothetical protein VITISV_030941 [Vitis vinifera] |
| 4 |
Hb_001958_010 |
0.0851335059 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 5 |
Hb_004076_060 |
0.0860980498 |
- |
- |
PREDICTED: UDP-glycosyltransferase 82A1 [Jatropha curcas] |
| 6 |
Hb_012655_010 |
0.0903866189 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 7 |
Hb_003894_170 |
0.0963943491 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 8 |
Hb_017043_040 |
0.0991729685 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 9 |
Hb_004037_040 |
0.1189278921 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 10 |
Hb_001348_040 |
0.1226476308 |
- |
- |
hypothetical protein VITISV_006651 [Vitis vinifera] |
| 11 |
Hb_162174_010 |
0.1227797888 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_002391_030 |
0.1236001322 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 13 |
Hb_001514_030 |
0.1242473134 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 14 |
Hb_001079_060 |
0.1271790903 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001516_010 |
0.1295180818 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 16 |
Hb_004122_010 |
0.1303860966 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 17 |
Hb_174168_010 |
0.1308589553 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 18 |
Hb_007520_050 |
0.131481813 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 19 |
Hb_000941_110 |
0.1317355304 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 20 |
Hb_032572_010 |
0.1338482961 |
- |
- |
PREDICTED: phospholipase A2-alpha [Jatropha curcas] |