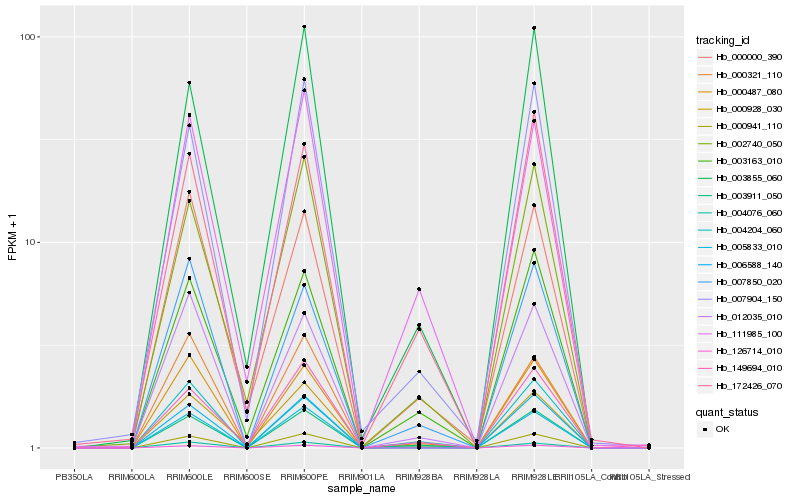
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003911_050 |
0.0 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 2 |
Hb_003163_010 |
0.074376885 |
- |
- |
hypothetical protein POPTR_0012s02920g [Populus trichocarpa] |
| 3 |
Hb_007904_150 |
0.0885259444 |
- |
- |
PREDICTED: probable pectate lyase 5 [Populus euphratica] |
| 4 |
Hb_172426_070 |
0.0933601476 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_007850_020 |
0.0934435157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_012035_010 |
0.0944657798 |
- |
- |
hypothetical protein JCGZ_21143 [Jatropha curcas] |
| 7 |
Hb_000941_110 |
0.0956639957 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 8 |
Hb_002740_050 |
0.0962830989 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86-like [Populus euphratica] |
| 9 |
Hb_004204_060 |
0.09646558 |
transcription factor |
TF Family: NAC |
NAC transcription factor 058, partial [Jatropha curcas] |
| 10 |
Hb_003855_060 |
0.099809386 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 11 |
Hb_126714_010 |
0.1013211591 |
- |
- |
hypothetical protein CISIN_1g041016mg, partial [Citrus sinensis] |
| 12 |
Hb_111985_100 |
0.1014972598 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 13 |
Hb_006588_140 |
0.101910779 |
- |
- |
PREDICTED: uncharacterized protein LOC104906772 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_004076_060 |
0.1027599818 |
- |
- |
PREDICTED: UDP-glycosyltransferase 82A1 [Jatropha curcas] |
| 15 |
Hb_149694_010 |
0.104932381 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_005833_010 |
0.1064678411 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000487_080 |
0.1094662833 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 18 |
Hb_000321_110 |
0.1101301868 |
- |
- |
Uncharacterized protein TCM_041937 [Theobroma cacao] |
| 19 |
Hb_000000_390 |
0.1134966086 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 20 |
Hb_000928_030 |
0.1142822004 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |