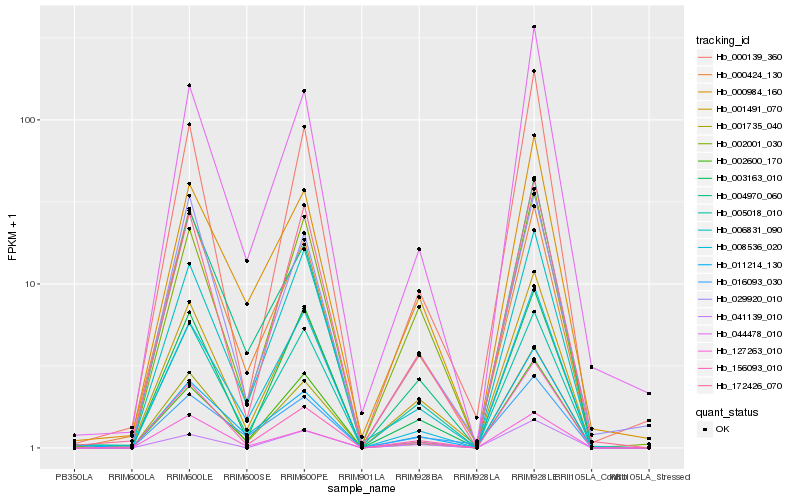
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001491_070 |
0.0 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 2 |
Hb_172426_070 |
0.0694311708 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_005018_010 |
0.0759583428 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 4 |
Hb_003163_010 |
0.0785971002 |
- |
- |
hypothetical protein POPTR_0012s02920g [Populus trichocarpa] |
| 5 |
Hb_002001_030 |
0.0793572356 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 6 |
Hb_000424_130 |
0.0909213271 |
- |
- |
PREDICTED: monoacylglycerol lipase abhd6-B-like [Jatropha curcas] |
| 7 |
Hb_000139_360 |
0.0912005888 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 8 |
Hb_001735_040 |
0.0973624719 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 9 |
Hb_008536_020 |
0.0975435485 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 10 |
Hb_006831_090 |
0.0997156299 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000984_160 |
0.1001848334 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_016093_030 |
0.1039254848 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 13 |
Hb_044478_010 |
0.1053655495 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 14 |
Hb_156093_010 |
0.1057221999 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 15 |
Hb_127263_010 |
0.1060512364 |
- |
- |
hypothetical protein JCGZ_02828 [Jatropha curcas] |
| 16 |
Hb_029920_010 |
0.1075403002 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 17 |
Hb_002600_170 |
0.1082099842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_041139_010 |
0.1086789613 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 19 |
Hb_004970_060 |
0.1090018602 |
- |
- |
PREDICTED: uncharacterized protein LOC100259061 [Vitis vinifera] |
| 20 |
Hb_011214_130 |
0.109339312 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 [Jatropha curcas] |