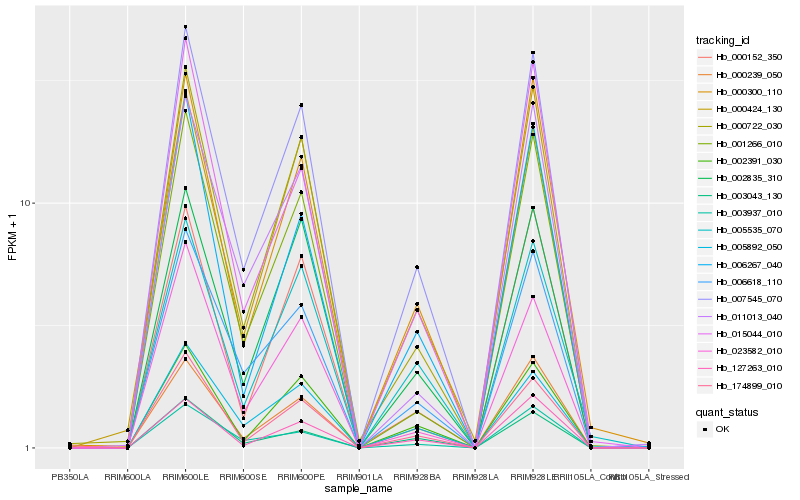
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007545_070 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 306 [Populus euphratica] |
| 2 |
Hb_006618_110 |
0.058837546 |
- |
- |
- |
| 3 |
Hb_000300_110 |
0.0733477929 |
- |
- |
hypothetical protein JCGZ_02651 [Jatropha curcas] |
| 4 |
Hb_005892_050 |
0.0749672025 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 5 |
Hb_000239_050 |
0.0750462739 |
- |
- |
PREDICTED: cyclin-D5-1-like [Jatropha curcas] |
| 6 |
Hb_002835_310 |
0.0760555576 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_000424_130 |
0.0781895293 |
- |
- |
PREDICTED: monoacylglycerol lipase abhd6-B-like [Jatropha curcas] |
| 8 |
Hb_002391_030 |
0.078571539 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 9 |
Hb_174899_010 |
0.0833553946 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 10 |
Hb_000722_030 |
0.0869512913 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 11 |
Hb_005535_070 |
0.0889210852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001266_010 |
0.0896801942 |
- |
- |
PREDICTED: uncharacterized protein LOC101206474 [Cucumis sativus] |
| 13 |
Hb_015044_010 |
0.0899827326 |
- |
- |
hypothetical protein JCGZ_25130 [Jatropha curcas] |
| 14 |
Hb_000152_350 |
0.0911371985 |
- |
- |
PREDICTED: enolase [Jatropha curcas] |
| 15 |
Hb_011013_040 |
0.0913708652 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 16 |
Hb_127263_010 |
0.0934861606 |
- |
- |
hypothetical protein JCGZ_02828 [Jatropha curcas] |
| 17 |
Hb_003937_010 |
0.0936708631 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 18 |
Hb_023582_010 |
0.0936743185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003043_130 |
0.0958395001 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 20 |
Hb_006267_040 |
0.098237182 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |