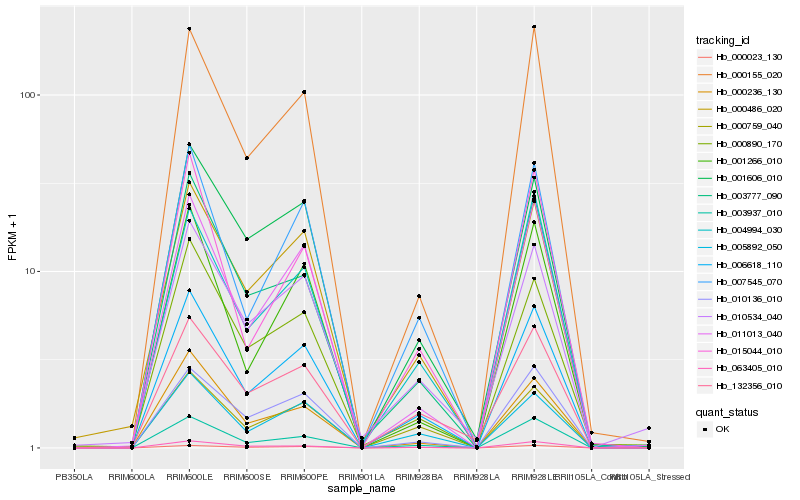
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006618_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_007545_070 |
0.058837546 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 306 [Populus euphratica] |
| 3 |
Hb_003937_010 |
0.062275541 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 4 |
Hb_005892_050 |
0.0710922603 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 5 |
Hb_000486_020 |
0.076638835 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 6 |
Hb_011013_040 |
0.0812686222 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 7 |
Hb_010136_010 |
0.0855707631 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 8 |
Hb_000759_040 |
0.0859033975 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 9 |
Hb_000155_020 |
0.0867741959 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_132356_010 |
0.0887298842 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 11 |
Hb_063405_010 |
0.0918435238 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 12 |
Hb_000236_130 |
0.0919235831 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 13 |
Hb_000023_130 |
0.093576549 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 14 |
Hb_003777_090 |
0.0937867681 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 15 |
Hb_004994_030 |
0.0940422485 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 16 |
Hb_001606_010 |
0.0959410196 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_000890_170 |
0.0990857654 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 18 |
Hb_010534_040 |
0.0992271376 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 19 |
Hb_015044_010 |
0.0998768805 |
- |
- |
hypothetical protein JCGZ_25130 [Jatropha curcas] |
| 20 |
Hb_001266_010 |
0.1012983648 |
- |
- |
PREDICTED: uncharacterized protein LOC101206474 [Cucumis sativus] |