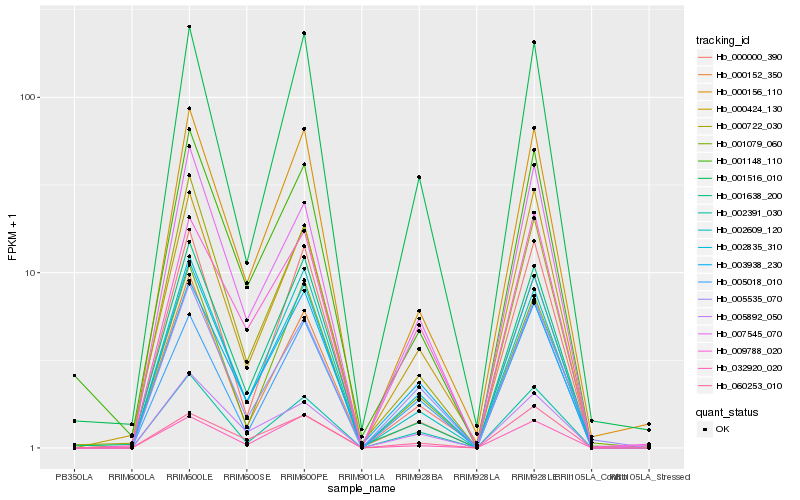
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002835_310 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_000156_110 |
0.0574465533 |
- |
- |
Uncharacterized protein TCM_029490 [Theobroma cacao] |
| 3 |
Hb_002391_030 |
0.0638405086 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 4 |
Hb_000424_130 |
0.0753717416 |
- |
- |
PREDICTED: monoacylglycerol lipase abhd6-B-like [Jatropha curcas] |
| 5 |
Hb_001516_010 |
0.0757524419 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 6 |
Hb_007545_070 |
0.0760555576 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 306 [Populus euphratica] |
| 7 |
Hb_003938_230 |
0.0776998174 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 8 |
Hb_002609_120 |
0.0783320196 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 9 |
Hb_000000_390 |
0.080988138 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 10 |
Hb_005535_070 |
0.0814621534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_032920_020 |
0.0840397145 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_001079_060 |
0.0902118472 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005018_010 |
0.0907320475 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 14 |
Hb_001638_200 |
0.0911847719 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 43 [Jatropha curcas] |
| 15 |
Hb_000722_030 |
0.0925751584 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 16 |
Hb_000152_350 |
0.0937948228 |
- |
- |
PREDICTED: enolase [Jatropha curcas] |
| 17 |
Hb_001148_110 |
0.0938737831 |
- |
- |
- |
| 18 |
Hb_060253_010 |
0.0955745111 |
- |
- |
hypothetical protein JCGZ_14707 [Jatropha curcas] |
| 19 |
Hb_005892_050 |
0.0973982609 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 20 |
Hb_009788_020 |
0.0977121228 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g06840 [Jatropha curcas] |