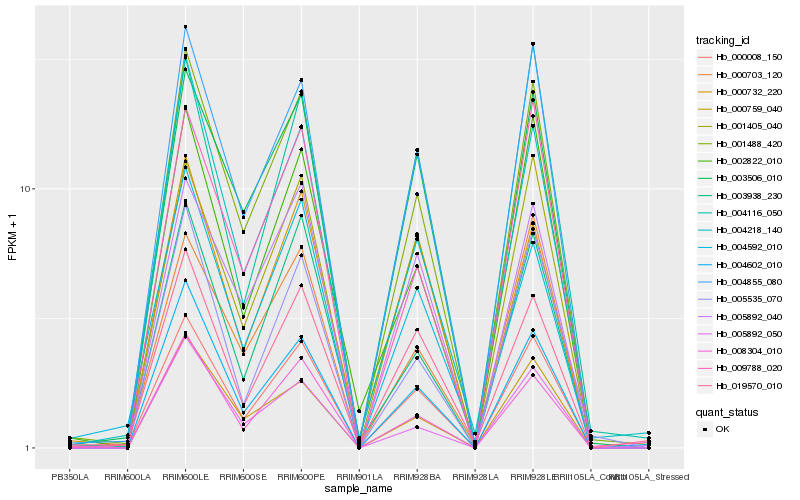
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_150 |
0.0 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 2 |
Hb_004855_080 |
0.0500505193 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 3 |
Hb_001488_420 |
0.0568948018 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 4 |
Hb_001405_040 |
0.0745511504 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 5 |
Hb_004602_010 |
0.0761242006 |
- |
- |
PREDICTED: uncharacterized protein LOC105042236 isoform X2 [Elaeis guineensis] |
| 6 |
Hb_003506_010 |
0.0801471073 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 7 |
Hb_005892_040 |
0.0907527102 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 8 |
Hb_000759_040 |
0.0925374848 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 9 |
Hb_008304_010 |
0.0953897933 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_009788_020 |
0.0954655197 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g06840 [Jatropha curcas] |
| 11 |
Hb_003938_230 |
0.0957853249 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 12 |
Hb_019570_010 |
0.0973226266 |
- |
- |
lyase, putative [Ricinus communis] |
| 13 |
Hb_002822_010 |
0.0986724583 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 14 |
Hb_004218_140 |
0.0996662309 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |
| 15 |
Hb_004116_050 |
0.1016022165 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 16 |
Hb_000703_120 |
0.1051885014 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000732_220 |
0.1076485248 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 18 |
Hb_004592_010 |
0.1084598632 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b-like [Jatropha curcas] |
| 19 |
Hb_005892_050 |
0.1098758804 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 20 |
Hb_005535_070 |
0.1102518246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |