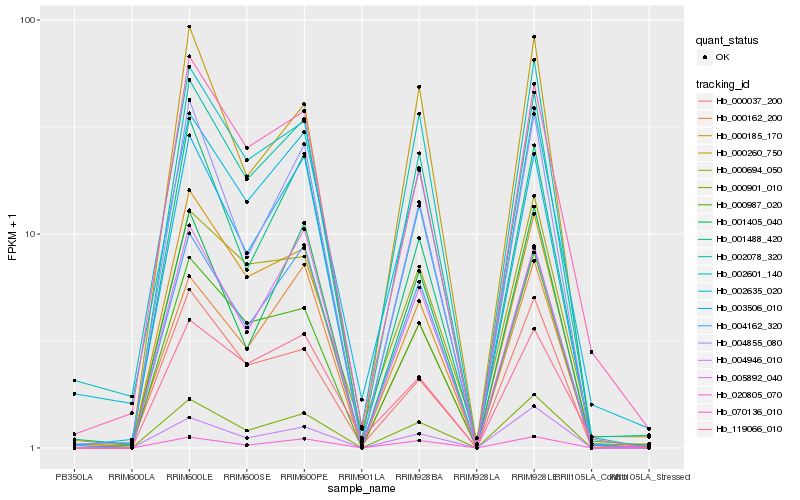
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_320 |
0.0 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 2 |
Hb_003506_010 |
0.0603998512 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 3 |
Hb_004162_320 |
0.0694402009 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000901_010 |
0.0703200673 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Jatropha curcas] |
| 5 |
Hb_000987_020 |
0.0729763098 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 6 |
Hb_000185_170 |
0.0763666045 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_020805_070 |
0.0864049863 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 8 |
Hb_004855_080 |
0.089732882 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 9 |
Hb_070136_010 |
0.0906655104 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 10 |
Hb_005892_040 |
0.0919295465 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 11 |
Hb_002601_140 |
0.0937521725 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 12 |
Hb_002635_020 |
0.093929581 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 13 |
Hb_000260_750 |
0.0982275256 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 14 |
Hb_001488_420 |
0.1015722504 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 15 |
Hb_000162_200 |
0.10304415 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000694_050 |
0.1040583193 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 17 |
Hb_001405_040 |
0.1063555835 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 18 |
Hb_000037_200 |
0.109943013 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_004946_010 |
0.1111947399 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 20 |
Hb_119066_010 |
0.1118255865 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |