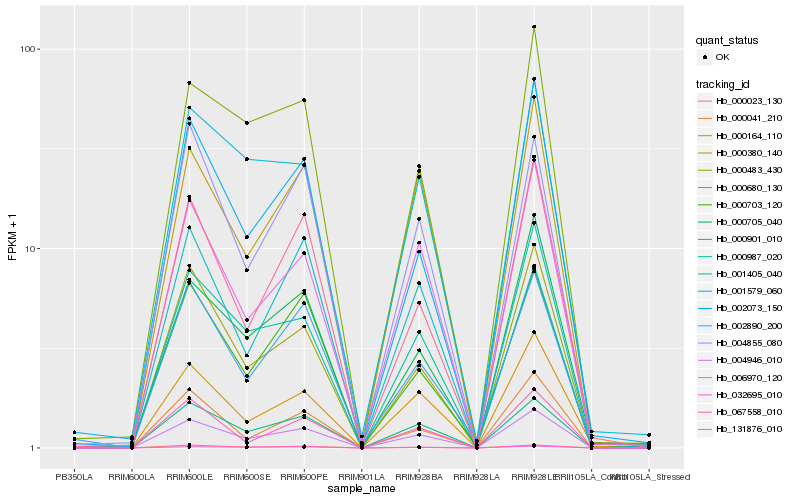
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004946_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 2 |
Hb_000901_010 |
0.0797745026 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Jatropha curcas] |
| 3 |
Hb_000380_140 |
0.0808870142 |
- |
- |
PREDICTED: uncharacterized protein LOC105628069 [Jatropha curcas] |
| 4 |
Hb_006970_120 |
0.0873317506 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000483_430 |
0.0888350528 |
- |
- |
- |
| 6 |
Hb_000987_020 |
0.0915554424 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 7 |
Hb_000023_130 |
0.0978121736 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 8 |
Hb_002073_150 |
0.1010050995 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 9 |
Hb_000680_130 |
0.1025794374 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 10 |
Hb_131876_010 |
0.1028330948 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_004855_080 |
0.1043876847 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 12 |
Hb_000703_120 |
0.1050951079 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002890_200 |
0.105816057 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 14 |
Hb_000705_040 |
0.10678829 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001579_060 |
0.1068904207 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 16 |
Hb_000164_110 |
0.1084218736 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 17 |
Hb_067558_010 |
0.1088194408 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 18 |
Hb_000041_210 |
0.1103527386 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 42 [Jatropha curcas] |
| 19 |
Hb_032695_010 |
0.1105465674 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 20 |
Hb_001405_040 |
0.1105901025 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |