| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
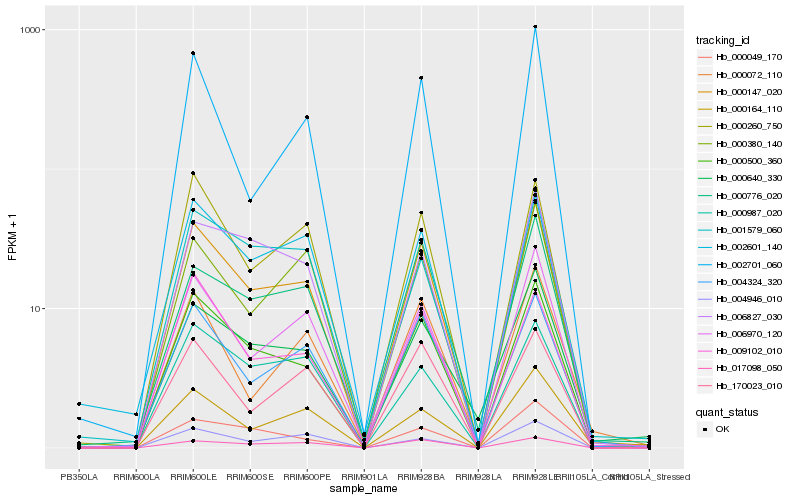
Hb_000147_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638303 [Jatropha curcas] |
| 2 |
Hb_000500_360 |
0.0585663207 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_006970_120 |
0.0850820253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004324_320 |
0.0879580551 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 5 |
Hb_000260_750 |
0.1068272059 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 6 |
Hb_000640_330 |
0.1073726845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000380_140 |
0.1141302722 |
- |
- |
PREDICTED: uncharacterized protein LOC105628069 [Jatropha curcas] |
| 8 |
Hb_170023_010 |
0.1186174395 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 9 |
Hb_002701_060 |
0.1188722129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002601_140 |
0.1191534792 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 11 |
Hb_009102_010 |
0.1192924882 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_017098_050 |
0.1199384844 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |
| 13 |
Hb_000164_110 |
0.1204317309 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 14 |
Hb_000776_020 |
0.1222267521 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 15 |
Hb_006827_030 |
0.1222941821 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 16 |
Hb_000049_170 |
0.122364065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001579_060 |
0.1235454919 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 18 |
Hb_004946_010 |
0.1259025564 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 19 |
Hb_000072_110 |
0.1263284716 |
- |
- |
PREDICTED: U-box domain-containing protein 35 [Jatropha curcas] |
| 20 |
Hb_000987_020 |
0.1279668875 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |