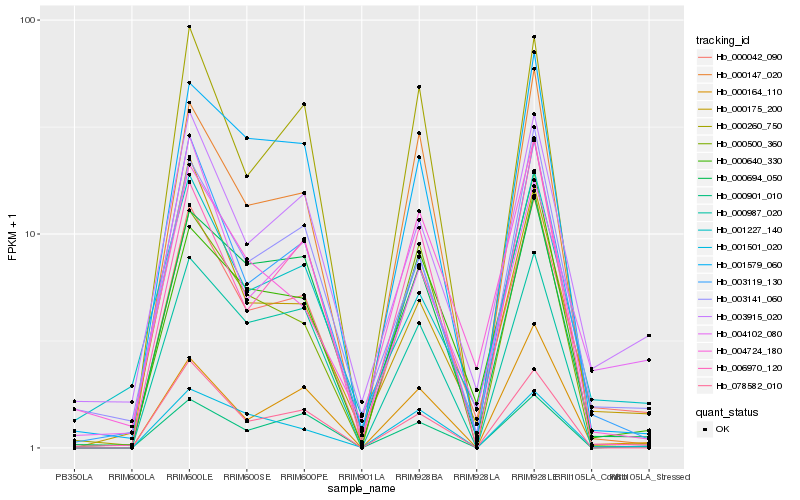
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_090 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 2 |
Hb_000640_330 |
0.1190907827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003915_020 |
0.1196850788 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003119_130 |
0.1285715131 |
- |
- |
hypothetical protein POPTR_0017s07770g [Populus trichocarpa] |
| 5 |
Hb_000164_110 |
0.1295776699 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 6 |
Hb_000147_020 |
0.1311953968 |
- |
- |
PREDICTED: uncharacterized protein LOC105638303 [Jatropha curcas] |
| 7 |
Hb_000694_050 |
0.1338967399 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 8 |
Hb_001501_020 |
0.1355873193 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 9 |
Hb_003141_060 |
0.1357761485 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 10 |
Hb_000260_750 |
0.1432820999 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 11 |
Hb_000500_360 |
0.1441407625 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000175_200 |
0.1453537194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004724_180 |
0.1479767111 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 14 |
Hb_000901_010 |
0.1480411155 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Jatropha curcas] |
| 15 |
Hb_001227_140 |
0.1483453894 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 16 |
Hb_004102_080 |
0.1493119435 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 17 |
Hb_006970_120 |
0.1506721847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001579_060 |
0.1511449708 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 19 |
Hb_078582_010 |
0.151449773 |
- |
- |
hypothetical protein POPTR_0005s01620g [Populus trichocarpa] |
| 20 |
Hb_000987_020 |
0.1522784967 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |