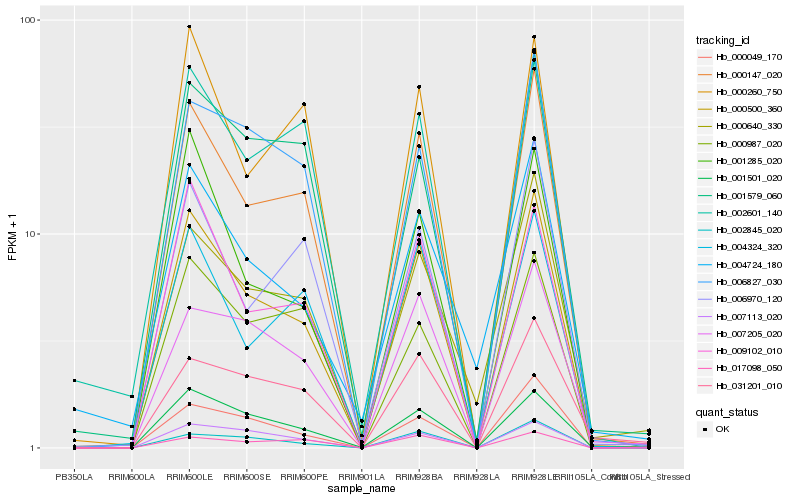
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000500_360 |
0.0 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000147_020 |
0.0585663207 |
- |
- |
PREDICTED: uncharacterized protein LOC105638303 [Jatropha curcas] |
| 3 |
Hb_001501_020 |
0.1010938786 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 4 |
Hb_009102_010 |
0.1023641449 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000049_170 |
0.1065818538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002845_020 |
0.113863121 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 7 |
Hb_001285_020 |
0.114608146 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_007113_020 |
0.1186847266 |
- |
- |
- |
| 9 |
Hb_004324_320 |
0.1221984084 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 10 |
Hb_007205_020 |
0.1229539894 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 11 |
Hb_006827_030 |
0.1261277921 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 12 |
Hb_000640_330 |
0.1281550314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000260_750 |
0.1300935389 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 14 |
Hb_006970_120 |
0.1338444078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001579_060 |
0.1386453217 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 16 |
Hb_002601_140 |
0.1400947748 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 17 |
Hb_031201_010 |
0.1406344521 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000987_020 |
0.1408389538 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 19 |
Hb_004724_180 |
0.1421058531 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 20 |
Hb_017098_050 |
0.1430301138 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |