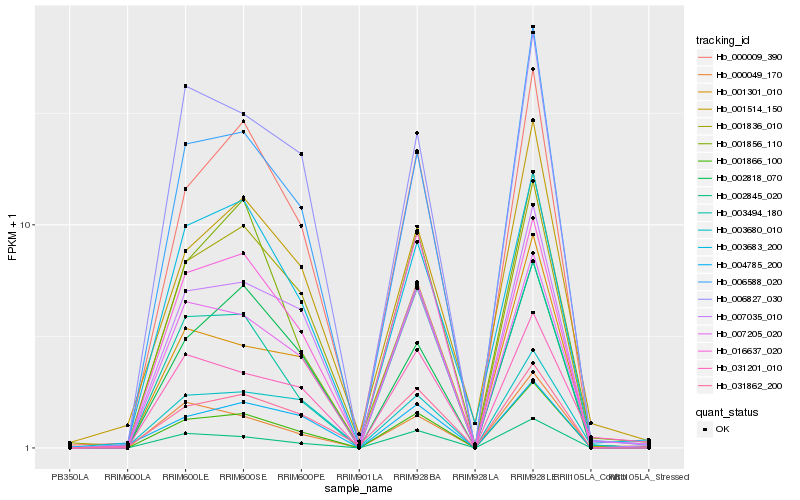
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001866_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 2 |
Hb_007035_010 |
0.0655896238 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000009_390 |
0.0709401388 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002845_020 |
0.0782390155 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 5 |
Hb_006588_020 |
0.0840411072 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_007205_020 |
0.0972192115 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 7 |
Hb_031862_200 |
0.1005419384 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 8 |
Hb_000049_170 |
0.1028625778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_016637_020 |
0.1069309595 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 10 |
Hb_006827_030 |
0.1069972784 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 11 |
Hb_003494_180 |
0.1098594013 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 12 |
Hb_001836_010 |
0.113166331 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 13 |
Hb_001301_010 |
0.1150797028 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 14 |
Hb_002818_070 |
0.1192292684 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 15 |
Hb_003683_200 |
0.1205762122 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 16 |
Hb_001514_150 |
0.1207556823 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 17 |
Hb_003680_010 |
0.1217556533 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_004785_200 |
0.1220605352 |
- |
- |
PREDICTED: protein MKS1 [Jatropha curcas] |
| 19 |
Hb_001856_110 |
0.1234156465 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |
| 20 |
Hb_031201_010 |
0.1335991984 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |