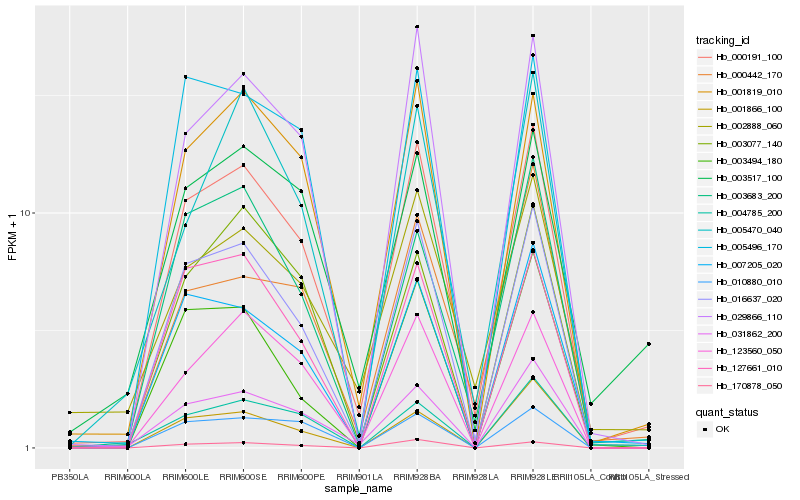
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000191_100 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_016637_020 |
0.0848432022 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 3 |
Hb_031862_200 |
0.0932375915 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 4 |
Hb_001819_010 |
0.0971803564 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 5 |
Hb_029866_110 |
0.098655439 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |
| 6 |
Hb_007205_020 |
0.1108775642 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 7 |
Hb_123560_050 |
0.114087134 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 8 |
Hb_170878_050 |
0.1200583395 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 9 |
Hb_002888_060 |
0.1221587953 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005496_170 |
0.1223581435 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 11 |
Hb_003683_200 |
0.127799615 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 12 |
Hb_127661_010 |
0.1296285415 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_003077_140 |
0.1330371246 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 14 |
Hb_010880_010 |
0.1333527905 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_005470_040 |
0.134561311 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 16 |
Hb_001866_100 |
0.1349665356 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 17 |
Hb_004785_200 |
0.1353109435 |
- |
- |
PREDICTED: protein MKS1 [Jatropha curcas] |
| 18 |
Hb_003494_180 |
0.1354760516 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 19 |
Hb_000442_170 |
0.1357264384 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 20 |
Hb_003517_100 |
0.1367104886 |
transcription factor |
TF Family: GRAS |
- |