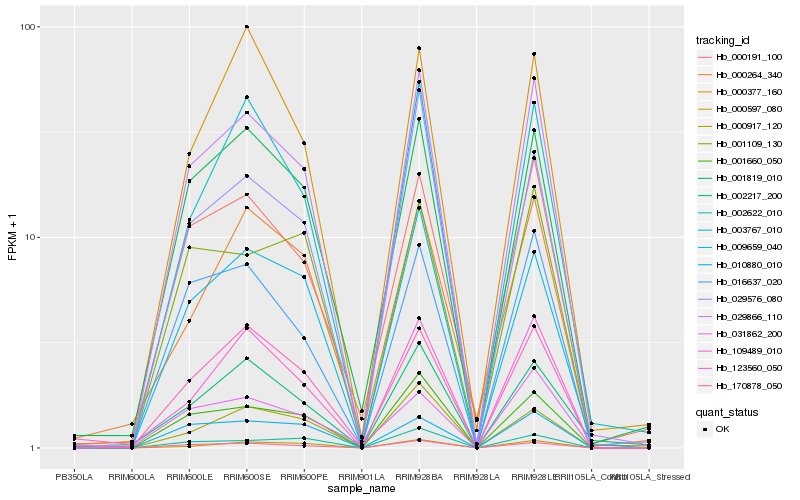
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029866_110 |
0.0 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |
| 2 |
Hb_170878_050 |
0.0705587098 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 3 |
Hb_001660_050 |
0.0769420803 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Eucalyptus grandis] |
| 4 |
Hb_123560_050 |
0.0930805415 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 5 |
Hb_016637_020 |
0.094025993 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 6 |
Hb_009659_040 |
0.0957262785 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 7 |
Hb_001819_010 |
0.0965274483 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 8 |
Hb_003767_010 |
0.0981964018 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 9 |
Hb_000191_100 |
0.098655439 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_029576_080 |
0.1104159242 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 11 |
Hb_000597_080 |
0.1127630133 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_109489_010 |
0.1185488397 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 13 |
Hb_000917_120 |
0.1192951424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002217_200 |
0.1227834334 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 15 |
Hb_010880_010 |
0.1242180313 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002622_010 |
0.124227021 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 17 |
Hb_001109_130 |
0.1256255404 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 136 isoform X1 [Jatropha curcas] |
| 18 |
Hb_031862_200 |
0.1277949013 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 19 |
Hb_000377_160 |
0.1328046986 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 20 |
Hb_000264_340 |
0.1346873239 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |