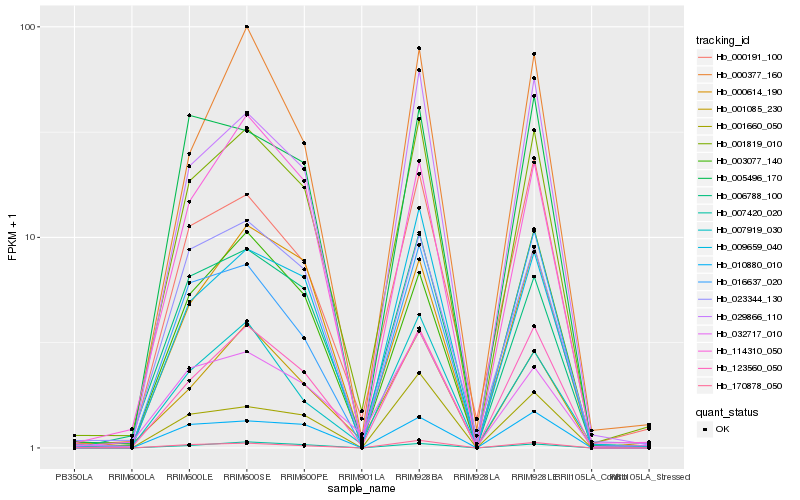
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001819_010 |
0.0 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 2 |
Hb_123560_050 |
0.0614210156 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 3 |
Hb_006788_100 |
0.0905876044 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 4 |
Hb_170878_050 |
0.0960539784 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 5 |
Hb_029866_110 |
0.0965274483 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |
| 6 |
Hb_000191_100 |
0.0971803564 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_009659_040 |
0.0991019184 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 8 |
Hb_001085_230 |
0.0996213154 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 9 |
Hb_023344_130 |
0.1000746062 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_007420_020 |
0.1065109002 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_010880_010 |
0.108416006 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003077_140 |
0.1092484861 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 13 |
Hb_016637_020 |
0.1127045879 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 14 |
Hb_000614_190 |
0.1136205393 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 15 |
Hb_114310_050 |
0.1165185154 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 16 |
Hb_000377_160 |
0.1167518894 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 17 |
Hb_032717_010 |
0.1186528455 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_005496_170 |
0.11939949 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 19 |
Hb_007919_030 |
0.1216149429 |
- |
- |
- |
| 20 |
Hb_001660_050 |
0.1216485733 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Eucalyptus grandis] |