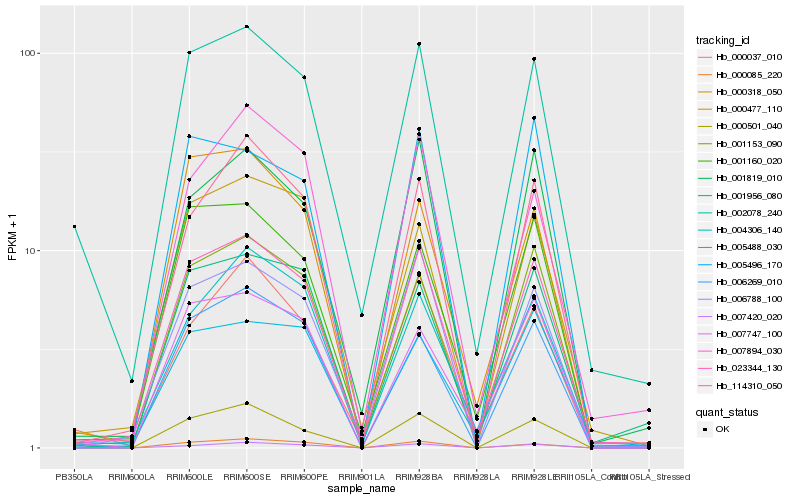
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023344_130 |
0.0 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_001153_090 |
0.0834056266 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 3 |
Hb_006788_100 |
0.0906994363 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001160_020 |
0.0977735297 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 5 |
Hb_002078_240 |
0.0985768297 |
rubber biosynthesis |
Gene Name: SRPP4 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 6 |
Hb_001956_080 |
0.098584874 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 7 |
Hb_000501_040 |
0.0990314698 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_001819_010 |
0.1000746062 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 9 |
Hb_114310_050 |
0.1026081805 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 10 |
Hb_000477_110 |
0.1053102044 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 11 |
Hb_004306_140 |
0.1077085113 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 12 |
Hb_007747_100 |
0.1098860536 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 13 |
Hb_007420_020 |
0.1106406371 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_005488_030 |
0.1109754415 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 15 |
Hb_000037_010 |
0.1122488897 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 16 |
Hb_000085_220 |
0.1127402482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006269_010 |
0.1131848222 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 18 |
Hb_000318_050 |
0.1197063619 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005496_170 |
0.1201583382 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 20 |
Hb_007894_030 |
0.1224032829 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |