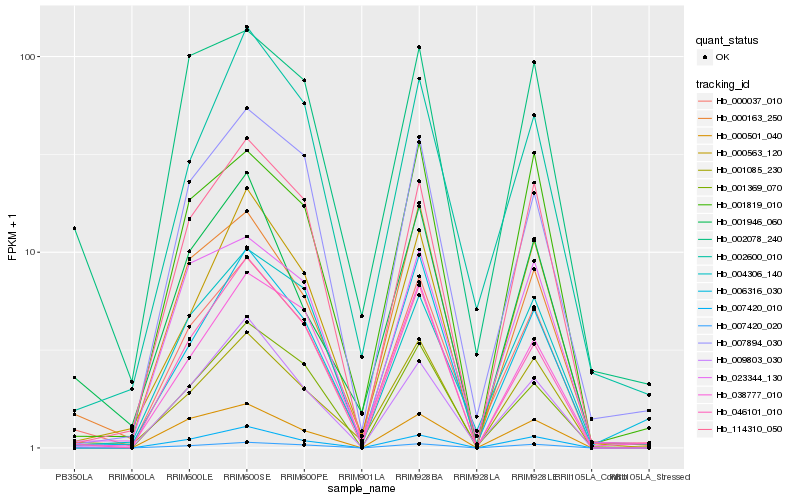
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000037_010 |
0.0 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 2 |
Hb_001085_230 |
0.098427665 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 3 |
Hb_009803_030 |
0.099662131 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 4 |
Hb_000163_250 |
0.1061848052 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_046101_010 |
0.1068811942 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 6 |
Hb_114310_050 |
0.1103772237 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 7 |
Hb_023344_130 |
0.1122488897 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_007420_020 |
0.1127302384 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 9 |
Hb_007420_010 |
0.1128767448 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001369_070 |
0.1166812656 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_007894_030 |
0.1203876434 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 12 |
Hb_004306_140 |
0.1223697934 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 13 |
Hb_000563_120 |
0.1230892857 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 14 |
Hb_002078_240 |
0.1241269678 |
rubber biosynthesis |
Gene Name: SRPP4 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 15 |
Hb_001946_060 |
0.1249672377 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002600_010 |
0.1259370018 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_001819_010 |
0.1264831994 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 18 |
Hb_038777_010 |
0.1265293464 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |
| 19 |
Hb_000501_040 |
0.127324156 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_006316_030 |
0.1276815017 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |