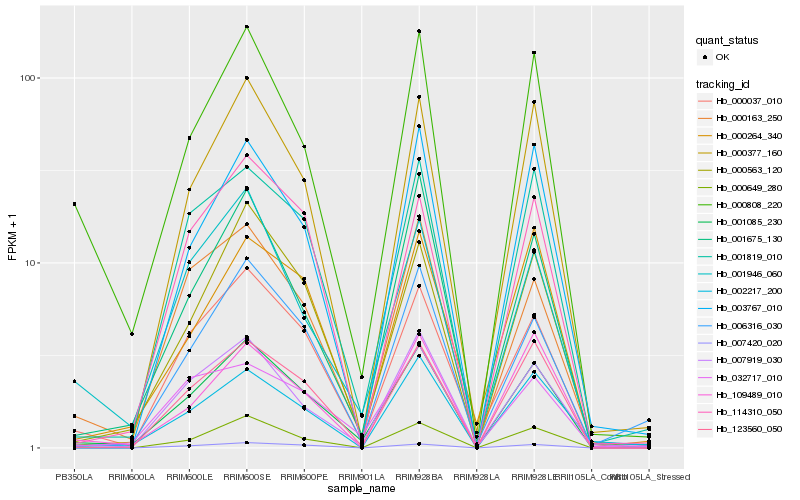
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_230 |
0.0 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 2 |
Hb_000037_010 |
0.098427665 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 3 |
Hb_001819_010 |
0.0996213154 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 4 |
Hb_123560_050 |
0.10319753 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 5 |
Hb_000563_120 |
0.1042074166 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 6 |
Hb_007919_030 |
0.1081012491 |
- |
- |
- |
| 7 |
Hb_000377_160 |
0.1119040734 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 8 |
Hb_000808_220 |
0.1142768352 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 9 |
Hb_000163_250 |
0.1179898322 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_001946_060 |
0.1195290059 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_006316_030 |
0.1206117572 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 12 |
Hb_000649_280 |
0.1221450407 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 13 |
Hb_114310_050 |
0.1282197254 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 14 |
Hb_001675_130 |
0.1291263809 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 15 |
Hb_032717_010 |
0.1318794897 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_002217_200 |
0.1324537535 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 17 |
Hb_003767_010 |
0.1330269865 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 18 |
Hb_007420_020 |
0.1342938335 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 19 |
Hb_109489_010 |
0.1342983834 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 20 |
Hb_000264_340 |
0.1351846647 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |