| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
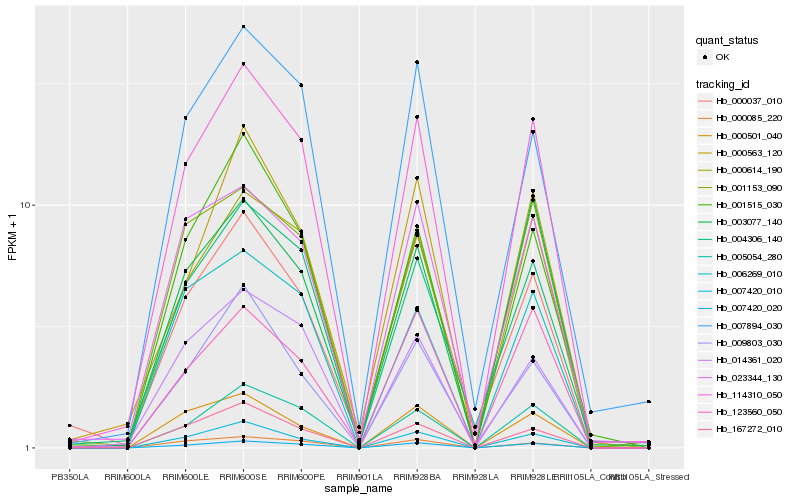
Hb_114310_050 |
0.0 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 2 |
Hb_007420_020 |
0.0562980163 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 3 |
Hb_005054_280 |
0.0692020458 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 4 |
Hb_000614_190 |
0.0702994179 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 5 |
Hb_007420_010 |
0.0761221622 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004306_140 |
0.0845413995 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 7 |
Hb_001515_030 |
0.0941349463 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_167272_010 |
0.0942774627 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 9 |
Hb_000563_120 |
0.0985311926 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 10 |
Hb_001153_090 |
0.1001363332 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 11 |
Hb_014361_020 |
0.1020560219 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 12 |
Hb_023344_130 |
0.1026081805 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_006269_010 |
0.1031556855 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 14 |
Hb_009803_030 |
0.1039140825 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 15 |
Hb_003077_140 |
0.1041484943 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 16 |
Hb_007894_030 |
0.1090334877 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 17 |
Hb_000501_040 |
0.109807933 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000037_010 |
0.1103772237 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 19 |
Hb_123560_050 |
0.1108238274 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 20 |
Hb_000085_220 |
0.1123134015 |
- |
- |
conserved hypothetical protein [Ricinus communis] |