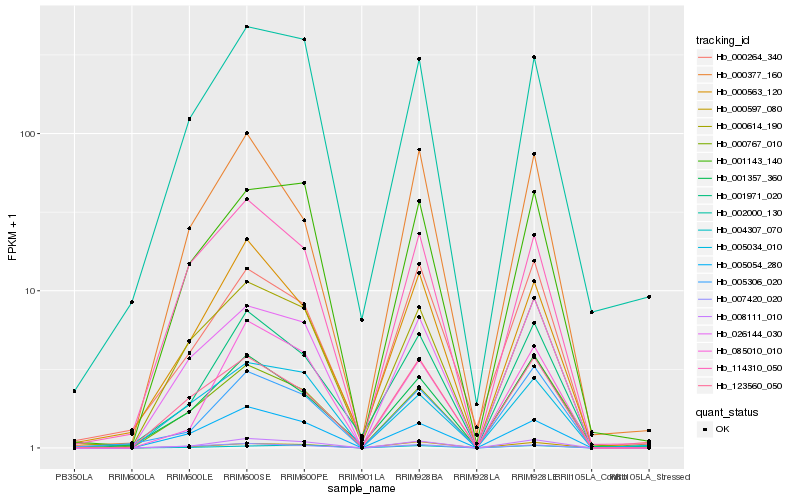
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008111_010 |
0.0 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 2 |
Hb_026144_030 |
0.0797044119 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 3 |
Hb_005054_280 |
0.0907407863 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 4 |
Hb_085010_010 |
0.0987517596 |
- |
- |
PREDICTED: MLO-like protein 3 isoform X2 [Vitis vinifera] |
| 5 |
Hb_005306_020 |
0.1003489921 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 6 |
Hb_001971_020 |
0.1010538878 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |
| 7 |
Hb_001143_140 |
0.1031813024 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000614_190 |
0.103516644 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 9 |
Hb_000597_080 |
0.1099222954 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000767_010 |
0.1103247108 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 11 |
Hb_007420_020 |
0.1137085509 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_001357_360 |
0.1137321344 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_123560_050 |
0.1209584214 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 14 |
Hb_000264_340 |
0.1288591956 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000563_120 |
0.1294043922 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 16 |
Hb_114310_050 |
0.1313271118 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 17 |
Hb_002000_130 |
0.1324570657 |
- |
- |
PREDICTED: abscisic stress-ripening protein 2-like [Eucalyptus grandis] |
| 18 |
Hb_005034_010 |
0.1325466178 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_004307_070 |
0.1345749845 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_000377_160 |
0.1368226733 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |