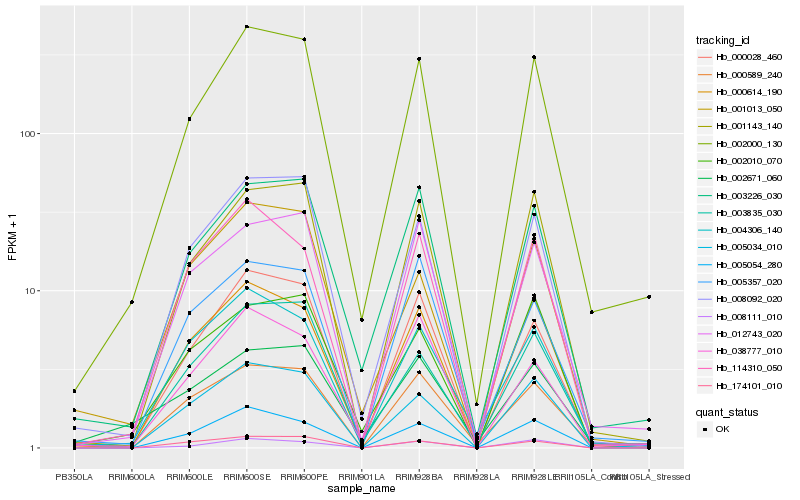
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002000_130 |
0.0 |
- |
- |
PREDICTED: abscisic stress-ripening protein 2-like [Eucalyptus grandis] |
| 2 |
Hb_003226_030 |
0.0844921461 |
- |
- |
sucrose synthase 5 [Hevea brasiliensis] |
| 3 |
Hb_008092_020 |
0.0905375133 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000028_460 |
0.0945821771 |
- |
- |
hypothetical protein POPTR_0015s05340g [Populus trichocarpa] |
| 5 |
Hb_003835_030 |
0.0951091191 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 6 |
Hb_001143_140 |
0.095893079 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000614_190 |
0.1008585719 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 8 |
Hb_012743_020 |
0.1011349614 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 9 |
Hb_005034_010 |
0.104961322 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_004306_140 |
0.1119914729 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 11 |
Hb_005054_280 |
0.1186398375 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 12 |
Hb_000589_240 |
0.1197680987 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 13 |
Hb_001013_050 |
0.1263934151 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_114310_050 |
0.1268574048 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 15 |
Hb_005357_020 |
0.1278261537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002010_070 |
0.129006657 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 17 |
Hb_002671_060 |
0.130949354 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 18 |
Hb_008111_010 |
0.1324570657 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 19 |
Hb_174101_010 |
0.1326019351 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 20 |
Hb_038777_010 |
0.1331110093 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |