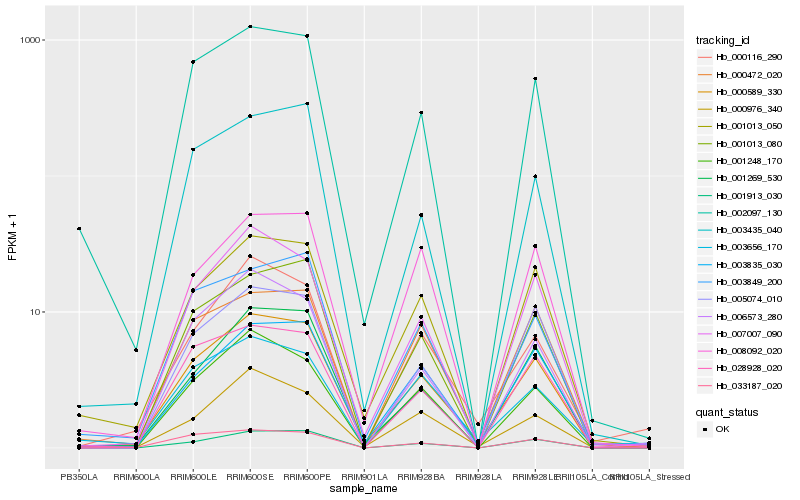
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000589_330 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_001013_050 |
0.0871399291 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_007007_090 |
0.0927686261 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 4 |
Hb_001913_030 |
0.095701867 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 5 |
Hb_003835_030 |
0.0978189253 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 6 |
Hb_002097_130 |
0.0982088569 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 7 |
Hb_001269_530 |
0.0989019852 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 8 |
Hb_001013_080 |
0.1004435535 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 9 |
Hb_028928_020 |
0.1051856045 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 10 |
Hb_003849_200 |
0.1055591355 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_001248_170 |
0.1082980551 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 12 |
Hb_000116_290 |
0.1106312143 |
- |
- |
PREDICTED: proton-coupled amino acid transporter 3 [Jatropha curcas] |
| 13 |
Hb_000976_340 |
0.114003763 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_008092_020 |
0.1174118969 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_006573_280 |
0.1182485224 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 16 |
Hb_033187_020 |
0.1192709916 |
- |
- |
hypothetical protein CARUB_v10004250mg, partial [Capsella rubella] |
| 17 |
Hb_005074_010 |
0.1196930422 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 18 |
Hb_000472_020 |
0.1211696226 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 19 |
Hb_003435_040 |
0.1222737035 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 20 |
Hb_003656_170 |
0.1230533386 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |