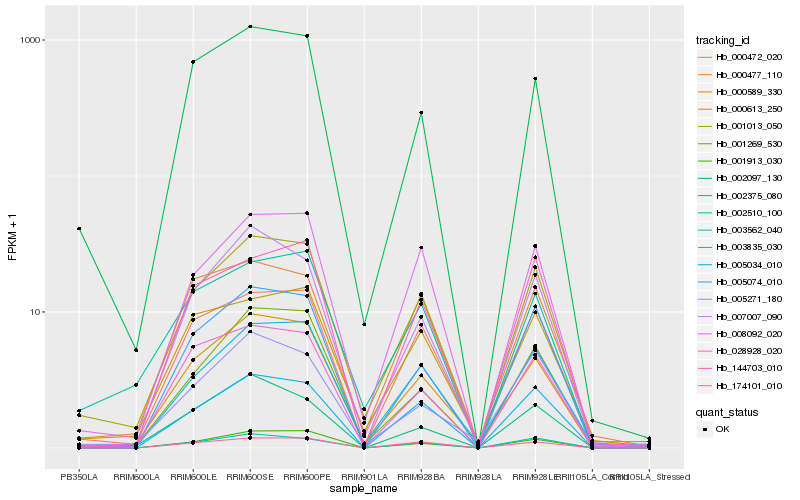
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001013_050 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_003835_030 |
0.0767807617 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 3 |
Hb_008092_020 |
0.0858240165 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000589_330 |
0.0871399291 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_000472_020 |
0.0932367011 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 6 |
Hb_000613_250 |
0.1025477634 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 7 |
Hb_002097_130 |
0.1042226591 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 8 |
Hb_001269_530 |
0.1080878372 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_007007_090 |
0.109836662 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 10 |
Hb_002510_100 |
0.1105469913 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 11 |
Hb_001913_030 |
0.1160643358 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 12 |
Hb_005074_010 |
0.1172282338 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 13 |
Hb_005034_010 |
0.1190119474 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_003562_040 |
0.1192408902 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 15 |
Hb_028928_020 |
0.11974838 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 16 |
Hb_005271_180 |
0.1200555979 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 17 |
Hb_174101_010 |
0.1200694693 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 18 |
Hb_000477_110 |
0.1216071394 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 19 |
Hb_144703_010 |
0.1220040161 |
- |
- |
Phospholipase A 2A, IIA,PLA2A [Theobroma cacao] |
| 20 |
Hb_002375_080 |
0.1245472177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |