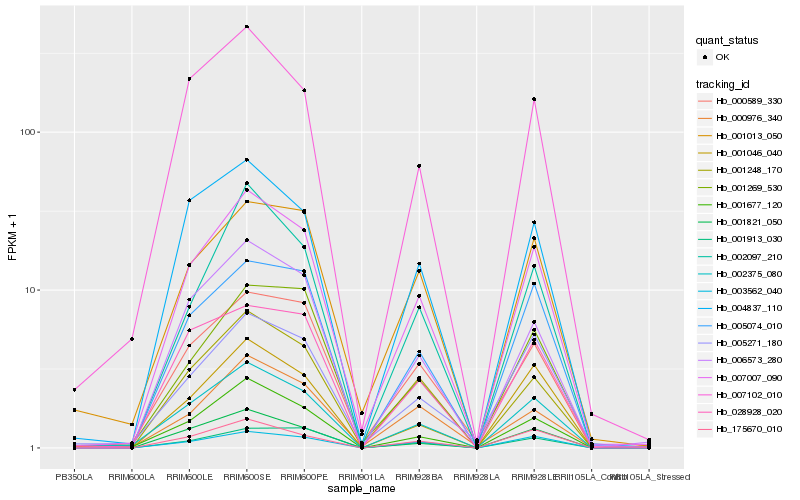
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007007_090 |
0.0 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 2 |
Hb_002375_080 |
0.0820928781 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001248_170 |
0.0883898455 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 4 |
Hb_001677_120 |
0.0913111053 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 5 |
Hb_000589_330 |
0.0927686261 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_006573_280 |
0.0979890601 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 7 |
Hb_003562_040 |
0.1012961732 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 8 |
Hb_005074_010 |
0.10236333 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 9 |
Hb_175670_010 |
0.1025735968 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 10 |
Hb_004837_110 |
0.1028117263 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001821_050 |
0.1076417698 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 12 |
Hb_005271_180 |
0.1094133936 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 13 |
Hb_001013_050 |
0.109836662 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_007102_010 |
0.1103628862 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 15 |
Hb_002097_210 |
0.1108989052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001046_040 |
0.1122832351 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_001269_530 |
0.1138687069 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 18 |
Hb_028928_020 |
0.1140776334 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 19 |
Hb_000976_340 |
0.1152580736 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_001913_030 |
0.1211572917 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |