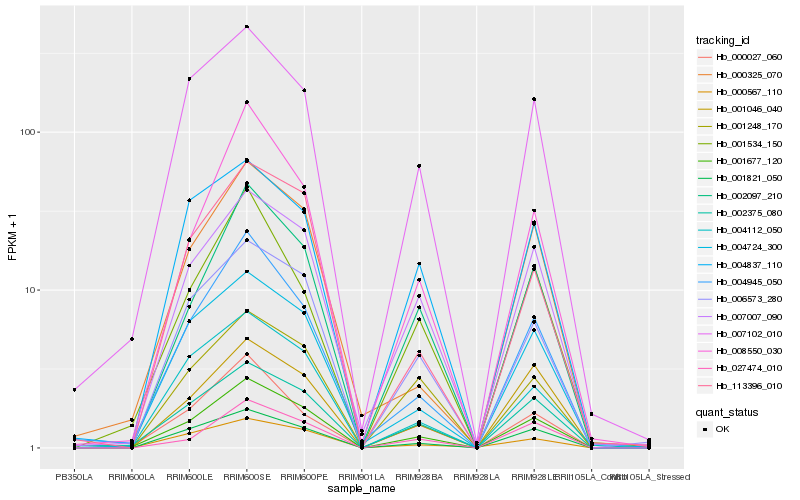
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001677_120 |
0.0 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 2 |
Hb_002375_080 |
0.0875852021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001821_050 |
0.0911117241 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 4 |
Hb_007007_090 |
0.0913111053 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 5 |
Hb_006573_280 |
0.0935766133 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 6 |
Hb_004724_300 |
0.0958846616 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 7 |
Hb_004945_050 |
0.0968324163 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 8 |
Hb_007102_010 |
0.0985250262 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 9 |
Hb_002097_210 |
0.1002966293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004112_050 |
0.1127658551 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_000325_070 |
0.1157316125 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_008550_030 |
0.1207615731 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |
| 13 |
Hb_001046_040 |
0.1217252345 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_027474_010 |
0.1245014304 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 15 |
Hb_113396_010 |
0.1247048304 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000027_060 |
0.1251065896 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 17 |
Hb_001248_170 |
0.1264942644 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 18 |
Hb_000567_110 |
0.1280662535 |
- |
- |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 19 |
Hb_004837_110 |
0.1307607726 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001534_150 |
0.130896686 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |