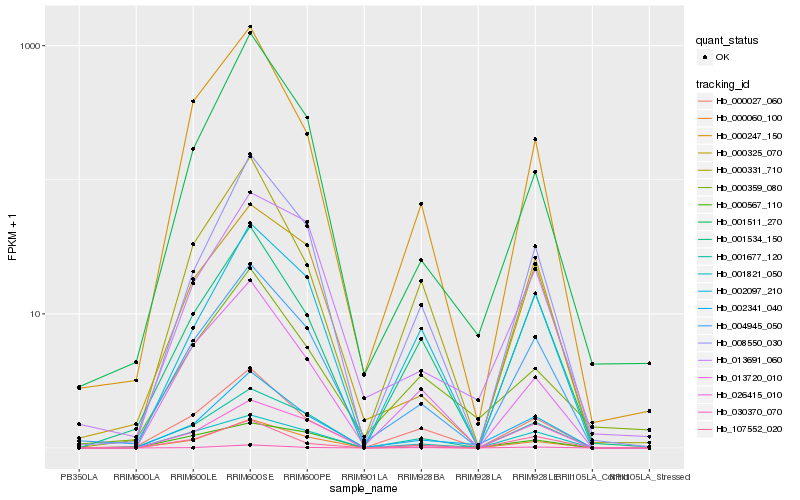
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002341_040 |
0.0 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 2 |
Hb_004945_050 |
0.1007208716 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 3 |
Hb_008550_030 |
0.1074849952 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |
| 4 |
Hb_000060_100 |
0.1286843216 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001677_120 |
0.1322445484 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 6 |
Hb_001534_150 |
0.1351644061 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 7 |
Hb_002097_210 |
0.1379886964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000027_060 |
0.1427728455 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 9 |
Hb_000325_070 |
0.1468181681 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_030370_070 |
0.1468674378 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_013691_060 |
0.1515234631 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 12 |
Hb_001511_270 |
0.1515366347 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 13 |
Hb_000247_150 |
0.1524778757 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 14 |
Hb_026415_010 |
0.1551807232 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000567_110 |
0.1566235193 |
- |
- |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 16 |
Hb_000331_710 |
0.1569093426 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_013720_010 |
0.1585160478 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |
| 18 |
Hb_107552_020 |
0.1588641973 |
- |
- |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 19 |
Hb_001821_050 |
0.1593471083 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 20 |
Hb_000359_080 |
0.1605742856 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |