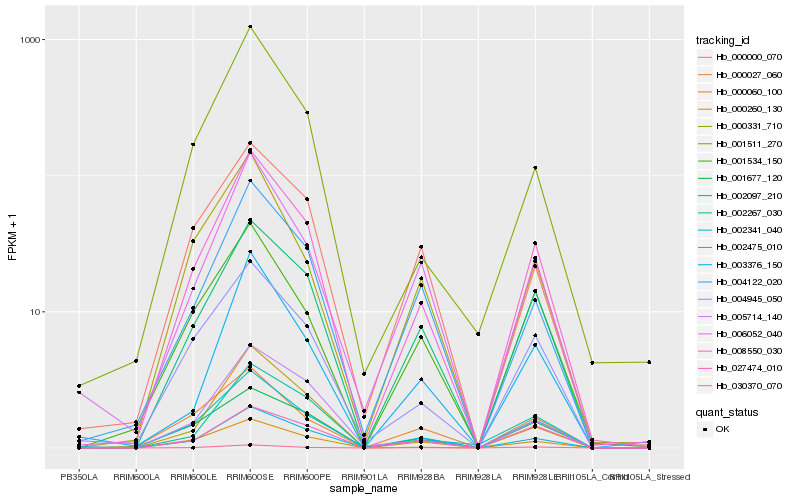
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008550_030 |
0.0 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |
| 2 |
Hb_002097_210 |
0.0881339165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004945_050 |
0.0978083171 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 4 |
Hb_002267_030 |
0.1054084709 |
- |
- |
hypothetical protein JCGZ_24883 [Jatropha curcas] |
| 5 |
Hb_002341_040 |
0.1074849952 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 6 |
Hb_030370_070 |
0.1146887583 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_000060_100 |
0.1176509089 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004122_020 |
0.1202659943 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 9 |
Hb_001677_120 |
0.1207615731 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 10 |
Hb_001534_150 |
0.1224869851 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 11 |
Hb_006052_040 |
0.1251795413 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 12 |
Hb_000027_060 |
0.1304046941 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 13 |
Hb_005714_140 |
0.1314865733 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000260_130 |
0.1317009083 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_002475_010 |
0.1341495293 |
- |
- |
hypothetical protein JCGZ_22530 [Jatropha curcas] |
| 16 |
Hb_001511_270 |
0.1344881466 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 17 |
Hb_003376_150 |
0.1351935115 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 18 |
Hb_027474_010 |
0.1352764406 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 19 |
Hb_000331_710 |
0.1353318322 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000000_070 |
0.142209956 |
- |
- |
allene oxide synthase [Hevea brasiliensis] |