| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
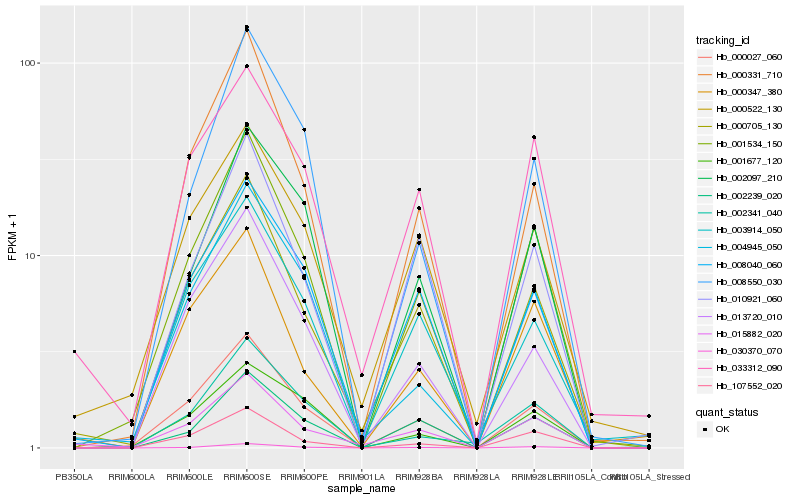
Hb_001534_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 2 |
Hb_000027_060 |
0.0667669711 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 3 |
Hb_030370_070 |
0.0943095236 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_107552_020 |
0.0996723252 |
- |
- |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 5 |
Hb_000705_130 |
0.1052950388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000331_710 |
0.106623851 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_008550_030 |
0.1224869851 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |
| 8 |
Hb_002097_210 |
0.1244022314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002239_020 |
0.1256118358 |
- |
- |
MLO, putative [Theobroma cacao] |
| 10 |
Hb_004945_050 |
0.1257808235 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 11 |
Hb_010921_060 |
0.1301178828 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_001677_120 |
0.130896686 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 13 |
Hb_015882_020 |
0.1311859922 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 14 |
Hb_008040_060 |
0.1326409538 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 15 |
Hb_033312_090 |
0.1344813176 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_003914_050 |
0.1347857295 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 17 |
Hb_002341_040 |
0.1351644061 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 18 |
Hb_000347_380 |
0.135706752 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_013720_010 |
0.1366166648 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |
| 20 |
Hb_000522_130 |
0.1369790238 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |