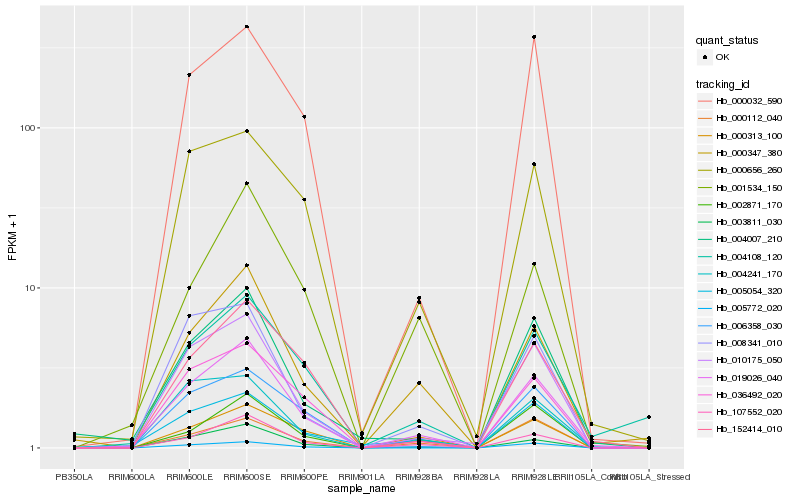
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019026_040 |
0.0 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 2 |
Hb_003811_030 |
0.1047137155 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 3 |
Hb_107552_020 |
0.1057669882 |
- |
- |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 4 |
Hb_010175_050 |
0.1149681778 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 5 |
Hb_000347_380 |
0.1262108775 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_036492_020 |
0.1304709837 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 7 |
Hb_004007_210 |
0.1320599503 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 8 |
Hb_152414_010 |
0.135428286 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein PRUPE_ppa024232mg [Prunus persica] |
| 9 |
Hb_005054_320 |
0.1396079981 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 10 |
Hb_000032_590 |
0.1421301003 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 11 |
Hb_000656_260 |
0.1452345126 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 12 |
Hb_004108_120 |
0.1479958286 |
- |
- |
PREDICTED: EID1-like F-box protein 3 [Jatropha curcas] |
| 13 |
Hb_006358_030 |
0.1484897417 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 14 |
Hb_002871_170 |
0.1488765229 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 15 |
Hb_004241_170 |
0.1508713478 |
- |
- |
PREDICTED: uncharacterized protein LOC105633220 [Jatropha curcas] |
| 16 |
Hb_008341_010 |
0.1509455529 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_005772_020 |
0.152863231 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 18 |
Hb_000112_040 |
0.157390666 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 19 |
Hb_001534_150 |
0.1575272486 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 20 |
Hb_000313_100 |
0.1576691548 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |