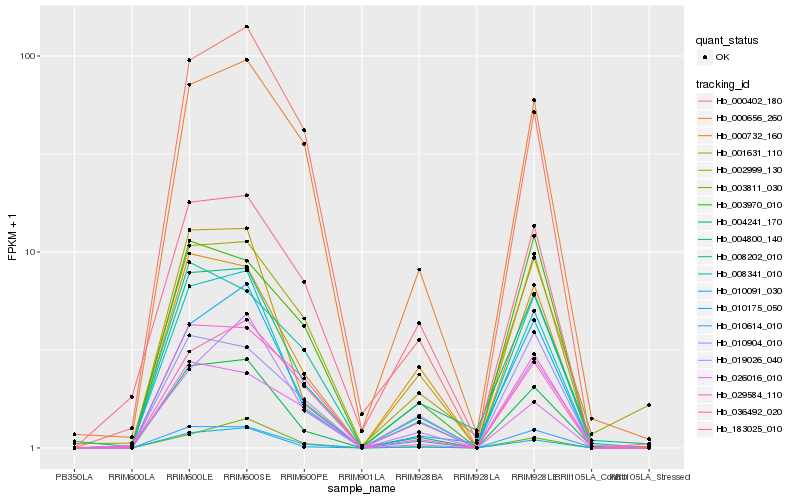
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004241_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633220 [Jatropha curcas] |
| 2 |
Hb_008341_010 |
0.0628364549 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001631_110 |
0.0895894869 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: gamma-glutamyltranspeptidase 1-like [Populus euphratica] |
| 4 |
Hb_010091_030 |
0.0992384794 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 5 |
Hb_004800_140 |
0.103119955 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000732_160 |
0.1064835243 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 7 |
Hb_010175_050 |
0.121228833 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 8 |
Hb_010614_010 |
0.1339478013 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 9 |
Hb_008202_010 |
0.1389904396 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_029584_110 |
0.1414861498 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 11 |
Hb_000656_260 |
0.1424101956 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 12 |
Hb_019026_040 |
0.1508713478 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 13 |
Hb_036492_020 |
0.1525037208 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 14 |
Hb_003811_030 |
0.1567594817 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 15 |
Hb_010904_010 |
0.1609358697 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000402_180 |
0.1632402098 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 17 |
Hb_183025_010 |
0.1647677891 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 18 |
Hb_002999_130 |
0.1660633489 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 19 |
Hb_026016_010 |
0.1697167498 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 20 |
Hb_003970_010 |
0.1698588091 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |