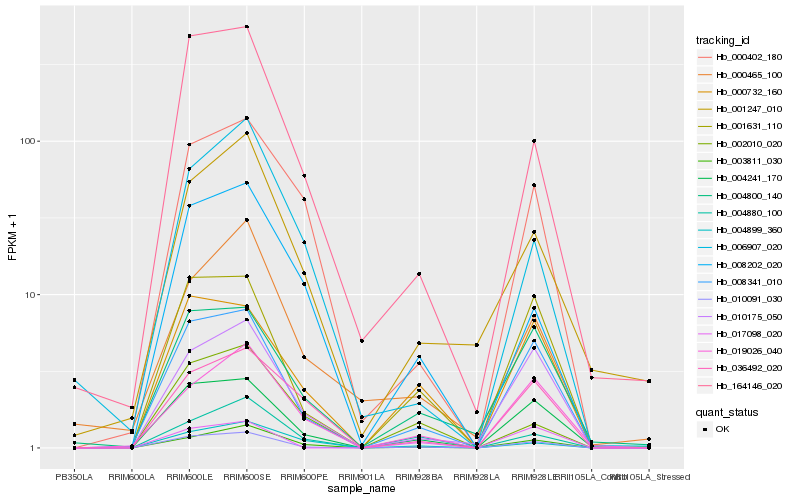
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010091_030 |
0.0 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 2 |
Hb_008341_010 |
0.078406201 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_004241_170 |
0.0992384794 |
- |
- |
PREDICTED: uncharacterized protein LOC105633220 [Jatropha curcas] |
| 4 |
Hb_001631_110 |
0.1180446079 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: gamma-glutamyltranspeptidase 1-like [Populus euphratica] |
| 5 |
Hb_003811_030 |
0.1363164614 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 6 |
Hb_010175_050 |
0.138795528 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 7 |
Hb_164146_020 |
0.1490551036 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_004800_140 |
0.1655529966 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000732_160 |
0.1725141675 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 10 |
Hb_019026_040 |
0.1756130179 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 11 |
Hb_000402_180 |
0.1821204212 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 12 |
Hb_008202_020 |
0.1886535144 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 13 |
Hb_001247_010 |
0.1887141604 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 14 |
Hb_036492_020 |
0.1896261821 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 15 |
Hb_004880_100 |
0.1902922668 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300-like [Glycine max] |
| 16 |
Hb_017098_020 |
0.1922139035 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 17 |
Hb_002010_020 |
0.1994931858 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 18 |
Hb_006907_020 |
0.1998271218 |
- |
- |
1-amino-cyclopropane-1-carboxylic acid oxidase 3 [Manihot esculenta] |
| 19 |
Hb_004899_360 |
0.2012084421 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 20 |
Hb_000465_100 |
0.2024747838 |
- |
- |
catalytic, putative [Ricinus communis] |