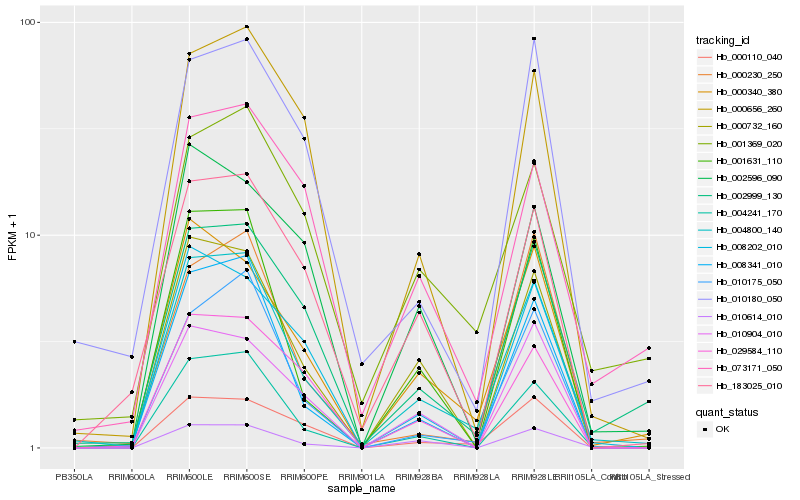
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004800_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004241_170 |
0.103119955 |
- |
- |
PREDICTED: uncharacterized protein LOC105633220 [Jatropha curcas] |
| 3 |
Hb_002999_130 |
0.1229471424 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 4 |
Hb_008341_010 |
0.1240467758 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000340_380 |
0.127139587 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 6 |
Hb_001631_110 |
0.1272567255 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: gamma-glutamyltranspeptidase 1-like [Populus euphratica] |
| 7 |
Hb_000732_160 |
0.1287413691 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 8 |
Hb_010175_050 |
0.1374232777 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 9 |
Hb_000110_040 |
0.140631961 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 10 |
Hb_010614_010 |
0.1432353531 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 11 |
Hb_000656_260 |
0.14333204 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 12 |
Hb_008202_010 |
0.1482905714 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_001369_020 |
0.149534755 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 14 |
Hb_010180_050 |
0.1504543573 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 15 |
Hb_183025_010 |
0.1524372985 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 16 |
Hb_000230_250 |
0.153479201 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_029584_110 |
0.1559144425 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 18 |
Hb_073171_050 |
0.1562923019 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 19 |
Hb_010904_010 |
0.1569579544 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002596_090 |
0.1643381988 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |