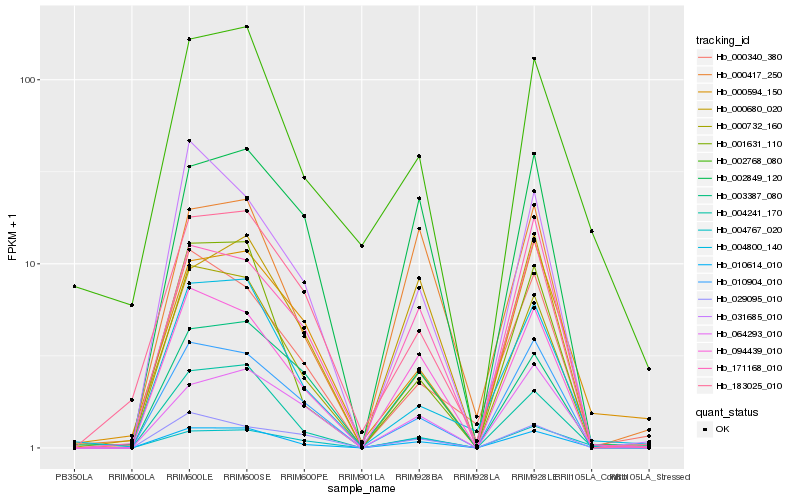
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010614_010 |
0.0 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 2 |
Hb_000732_160 |
0.0931118822 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 3 |
Hb_094439_010 |
0.1080999431 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 4 |
Hb_010904_010 |
0.1314482331 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004241_170 |
0.1339478013 |
- |
- |
PREDICTED: uncharacterized protein LOC105633220 [Jatropha curcas] |
| 6 |
Hb_001631_110 |
0.1356358433 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: gamma-glutamyltranspeptidase 1-like [Populus euphratica] |
| 7 |
Hb_004767_020 |
0.1415336044 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 8 |
Hb_004800_140 |
0.1432353531 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 9 |
Hb_064293_010 |
0.1447940818 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 10 |
Hb_171168_010 |
0.1531973446 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_031685_010 |
0.1540133301 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_002768_080 |
0.1601641514 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 13 |
Hb_002849_120 |
0.1618607286 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 14 |
Hb_000340_380 |
0.1638106257 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 15 |
Hb_003387_080 |
0.1639959143 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_183025_010 |
0.1640983478 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 17 |
Hb_000594_150 |
0.1657963694 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 18 |
Hb_000417_250 |
0.1661040569 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 19 |
Hb_029095_010 |
0.1661342193 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 20 |
Hb_000680_020 |
0.1663416032 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |