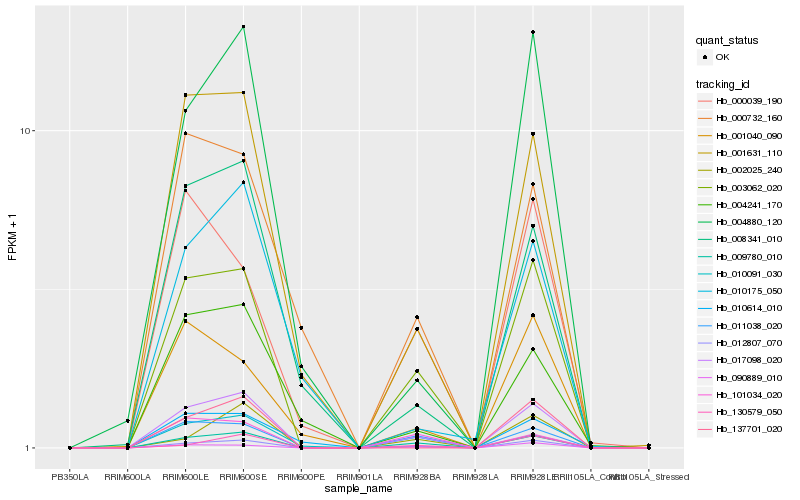
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017098_020 |
0.0 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 2 |
Hb_012807_070 |
0.0634999864 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 3 |
Hb_009780_010 |
0.0712867629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003062_020 |
0.0737511986 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 5 |
Hb_001631_110 |
0.1348048921 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: gamma-glutamyltranspeptidase 1-like [Populus euphratica] |
| 6 |
Hb_011038_020 |
0.1389417836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_090889_010 |
0.159346238 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 8 |
Hb_004880_120 |
0.174087584 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 9 |
Hb_008341_010 |
0.1753198279 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_010614_010 |
0.188551805 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 11 |
Hb_010091_030 |
0.1922139035 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 12 |
Hb_004241_170 |
0.1933511655 |
- |
- |
PREDICTED: uncharacterized protein LOC105633220 [Jatropha curcas] |
| 13 |
Hb_137701_020 |
0.1936961708 |
- |
- |
putative protein kinase [Oryza sativa Japonica Group] |
| 14 |
Hb_101034_020 |
0.1958455503 |
- |
- |
hypothetical protein JCGZ_13470 [Jatropha curcas] |
| 15 |
Hb_000732_160 |
0.198014161 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 16 |
Hb_002025_240 |
0.199239218 |
- |
- |
- |
| 17 |
Hb_130579_050 |
0.2035475431 |
- |
- |
hypothetical protein JCGZ_25577 [Jatropha curcas] |
| 18 |
Hb_000039_190 |
0.2101044959 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 19 |
Hb_010175_050 |
0.2102664388 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 20 |
Hb_001040_090 |
0.2105280331 |
- |
- |
PREDICTED: tetraspanin-19-like [Jatropha curcas] |